Hi MRtrix experts,
Using the recommended pipeline for multi-shell FBA produced a couple of strange artefacts in wmfod.mif when performing dwi2fod.
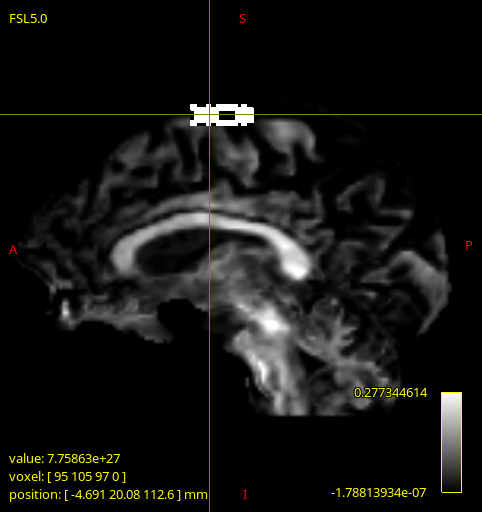
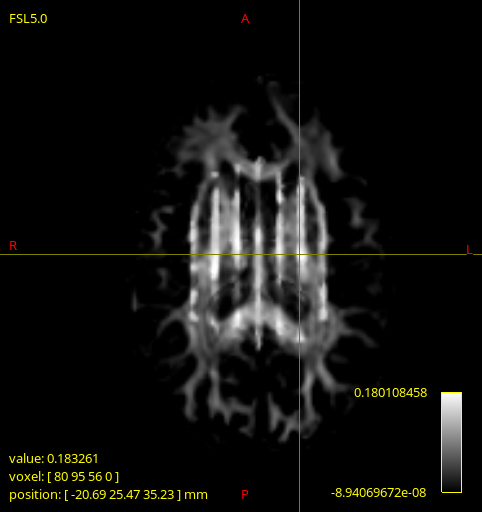
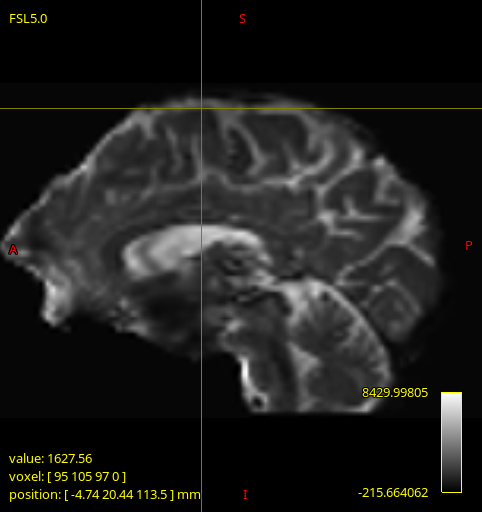
These artifacts (as seen below) only appeared on 2 of the DWI images being processed, and are quite different to each other. Strangely, upon inspecting the input image dwi_denoised_preproc_bias.mif there seems to be nothing wrong (though I could have missed something). The same artefacts are present in the corresponding gm.mif and csf.mif outputs too.
The commandline I used to run dwi2fod was as follows:
dwi2fod msmt_csd dwi_denoised_preproc_bias.mif ../group_average_response_wm.txt wmfod.mif ../group_average_response_gm.txt gm.mif ../group_average_response_csf.txt csf.mif -mask dwi_mask.mif -force
If you would like the image data to reproduce the error let me know.
Help much appreciated as always,
Matt
dwi2fod processed image1
dwi2fod processed image2
dwi_denoised_preproc_bias1.mif
dwi_denoised_preproc_bias2.mif