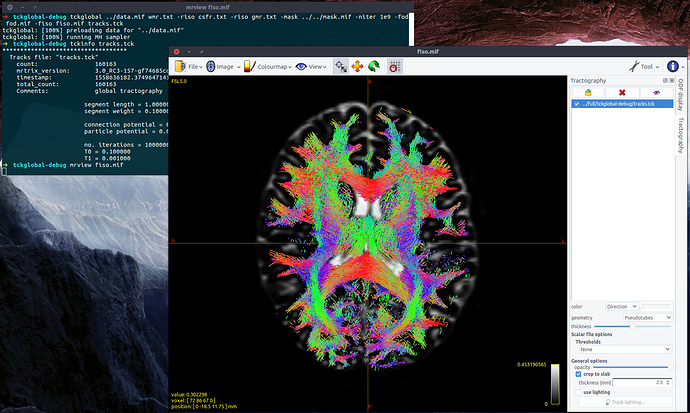
Hi to all MrTrixers!
I was interested in trying global tractography. We work on HCP data (multi-shell), and for other works we followed step-by-step the pipeline for MSMT-CSD as in the website. I launched global tracgtography following the suggested usage:
**tckglobal dwi.mif RF_WM.txt -riso RF_CSF.txt -riso RF_GM.txt -mask mymask.mif -niter 1e9 -fod fod.mif -fiso fiso.mif tracks.tck -nthreads 1 **
using tissue response functions as obtained from dwi2response msmt_5tt, all as default.
dwi2response msmt_5tt DWI.mif 5TT.mif RF_WM.txt RF_GM.txt RF_CSF.txt -voxels RF_voxels.mif
nthreads was set to 1 since my OS does not support multi-threading.
Tractogram results in a few (three or four) scattered streamlines, like these:
I also tried by increasing iterations and decreasing particle weights accordingly, but without any result.
Can anyone please help me? I can’t just figure out what can be wrong. Should I repeat the dwi2response step with different parameters? What should I check?
Thanks in advance
Gianpaolo
Hi Gianpaolo
Given that you’re working with HCP data, I would expect the default parameters as per the tutorial to work out of the box. We used HCP data in the paper too.
Have you checked that your tissue response functions are OK? When using msmt_5tt it is essential that your DWI.mif and T1-derived 5tt.mif are co-registered. Use the mrview overlay tool to verify that this is the case, then check your WM response with shview -response to make sure that it has the appropriate pancake shape.
If your response functions are fine, then the first global tractography parameters to play with would be the particle potential -ppot. You can think of this parameter as the cost of adding one particle. Given that barely any are present in your picture, you may need to reduce this. However, as said, I would expect the default to work well for HCP data.
If you can’t sort it out, can you please send me the HCP Subject ID and your tissue response functions?
Cheers
Daan
Hi Daan! Thanks a lot for your help!
Yes, DWI.mif and 5tt.mif are co-registered and WM response looks fine, here they are:
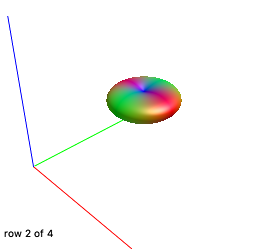
As expected, since we used them for other works (by using IFod2 tractography instead) and without any problem.
I’m not quite of an expert (sorry for my “noobness”) and I can’t explain myself why default settings doesn’t work properly. As you said, you also used HCP data in your paper.
Anyway, I’m currently tryng with decreasing -ppot and see what happens (it can’t go anyway worse than now  ) Do you suggest a factor by which I should decrease my -ppot to obtain some results?
) Do you suggest a factor by which I should decrease my -ppot to obtain some results?
HCP subject is 100307 anyway. I can send you RFs if you want.
Thank you again
Gianpaolo
Hi Daan,
I tried increasing -ppot as you suggested, but it didn’t showed any visible effect. It results in very sparse tracts as before. Can I write you privately and send you the estimated response functions?
Thanks for your kind help
Gianpaolo
Hi Gianpaolo,
Sorry for the late reply – I was away for ISMRM. Yes, please do send me your response functions and also the version of the code you’re using (tckglobal -version). I would like to get to the bottom of this.
Cheers
Daan
Hi Gianpaolo,
Thanks for sending me your response functions – they are perfectly fine. I was able to replicate the issue you reported when running the version from April 2017 that you were using. However, when I run the current master version of tckglobal all is fine (using the default parameters, HCP data and the response functions you sent me):
The only changes to tckglobal since 2017 have been to sort out an issue with multi-threading on some systems. I notice that you were using the -nthreads 1 flag; it’s good to know that in spite of this there was still a persistent issue. I would recommend updating your mrtrix install to a more recent version (git pull && ./build). This should not only fix the issue you were having, but also allow you to use multi-threading.
Hope this helps.
Daan

 ) Do you suggest a factor by which I should decrease my -ppot to obtain some results?
) Do you suggest a factor by which I should decrease my -ppot to obtain some results?