Hi, Experts,
My aim is to compare the fiber and FA values among different individual. For the fibers of each individual are quite different, I wanna to register the fibers to MNI152 space. At the same time, I wanna to preserve the FA values of each fiber.
As far as I can think, there’re two ways to convert brain fiber tracts to template space: 1) Generate Fiber tracts and the corresponding diffusion attributes (such as FA, MD…) of each fiber, then warp the fibers to the template space. 2) register T1 and FOD data (or DWI data in general) to template space, then generate the fiber tracts and the corresponding diffusion attributes.
1 . About warp tck file to template space.
The previous topic is here. I find it that the warp image of data1 does not cover the whole brain tracts. It is the same with “synantsInverseWarp.nii” which is created by ANTS.
ANTS 3 -m CC[MNI152_T1_1mm.nii, t1_mpr_ns_sag_iso.nii.gz, 1, 5] -t SyN[0.5] -r Gauss[2,0] -o t1_disease_liuyan_to_MNI_synants.nii -i 30x90x20 --use-Histogram-Matching
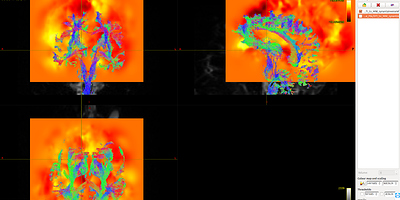
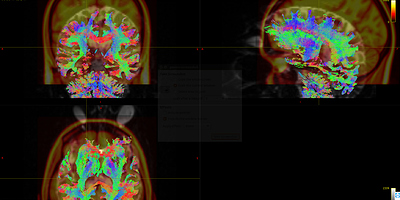
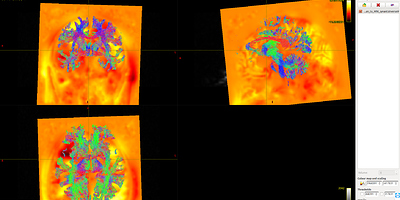
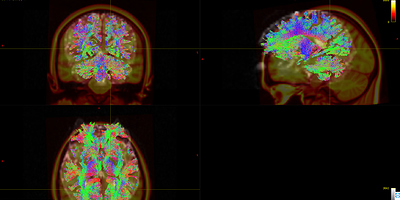
As shown in Fig 8.1-8.4, compared with data1, the whole brain tracts of data3 are covered by warp image. So, I should say the problem list on the first command ANTS. Does it mean the parameters of the ANTS is not suitable for the data? Is it the same to warp FA image as warp the fiber tracts?
2 . About register diffusion MRI to template space.
I learned that spatial normalization of diffusion data is not recommended, and never transform FOD data (or DWI data in general) .
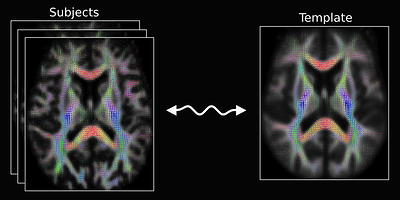
However, I find an Image that shows the registration for diffusion MRI, (see Fig 9): Designed explicitly for inter-subject alignment of fibre orientation distributions as provided by CSD and variants.
It seems that I should be able to achieve my goal through the above two methods. So, I am confused now.
Many Thanks,
Chaoqing
Fig 8.1 ( DTI_2013_act_0.5M.tck && T1_to_MNI_synantsInverseWarp ) – Data1
Fig 8.2 ( DTI_2013_act_0.5M.tck && MNI152_T1_1mm.nii.gz ) – Data1
Fig 8.3 ( DTI_30_average-6_act_sift_0.2M.tck && t1_disease_liuyan_to_MNI_synantsInverseWarp.ni ) – Data3
Fig 8.4 ( DTI_30_average-6_act_sift_0.2M.tck && MNI152_T1_1mm.nii.gz) – Data3
Fig 9.( Comes from MRtrix3 homepage: “Image registration for diffusion MRI” )