Hi again @Kar,
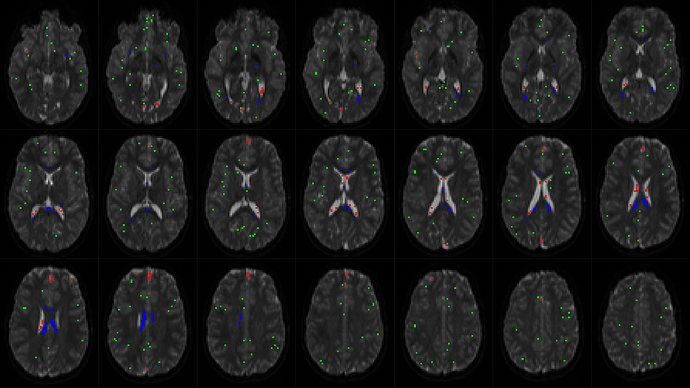
Looks generally ok. The only risk/concern in this case, which I can’t tell immediately from these images only, would be that your CSF response comes from an area that is “too generous”. You can confirm whether this is the case (or not) by looking at the -voxels output of dwi2response dhollander, and overlaying it on any of your images for reference. To do this, open any of your images (e.g. your actual “CSF” image you’ve shown above) in mrview, and open the voxel.mif (or whatever you named it) in the overlay tool. Next, in the overlay tool, switch “interpolation” off (at the bottom of the tool) and add a lower threshold of a value smaller than 1 (e.g. 0.1 or 0.5 or whatever  ). Then going to e.g. lightbox mode, you can generate a view that looks something like this:
). Then going to e.g. lightbox mode, you can generate a view that looks something like this:
This gives you/us a good idea of what voxels where exactly selected, so you can confirm if things make sense. Even though you’re only after WM-CSF CSD, it’s still quite a valuable quality-check to even pay close attention to the selected “GM” voxels (see my email). Working with rodent data, or for other people that may be working with ex-vivo data, or other funny/challenging scenarios, it may be worthwhile playing around with the -fa parameter to dwi2response dhollander. In any more “normal” scenario, this should never be needed. But in any “special” scenario, this may make the difference between things seemingly refusing to work and suddenly magically working out. In most of those “special” scenarios, the change to -fa you’d be looking for, is lowering its default value of 0.2 to e.g. 0.1. See again my email for details.
If you’re keen to share, feel free to post a few extra screenshots showing the voxel selection!
Cheers,
Thijs