Hello MRtrix experts,
I’m trying to use ACT, but for some reason streamlines in the corpus callosum seem to be rejected. I’m using single-shell data, b = 1000, voxel size = 2 mm isotropic, 64 directions, acquired in both AP and PA phase encoding directions. A Siemens Prisma machine was used to acquire the data.
Correction for EPI distortions, eddy currents and motion artefacts was done using FSL topup/eddy. The diffusion data was then aligned to a T1 scan using dtiInit function from the Vistasoft/MrDiffusion package.
The following commands were used MRtrix3 to track streamlines using ACT:
dwi2response tournier dwi.mif response.txt
dwi2fod csd dwi.mif response.txt fod.mif -mask dwi_mask.mif
5ttgen fsl T1.nii.gz 5tt.mif
tckgen fod.mif ACT.tck -act 5tt.mif -seed_image dwi_mask.mif -select 500000
This gave the following results:
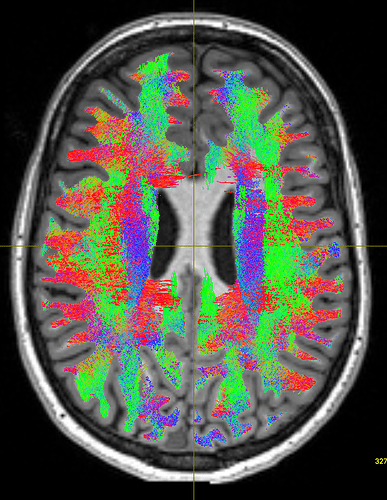
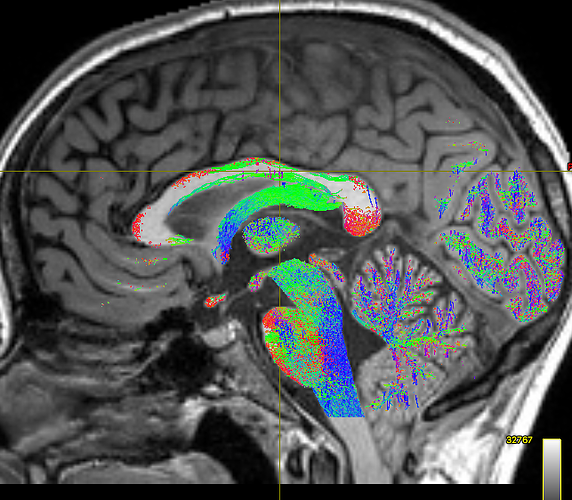
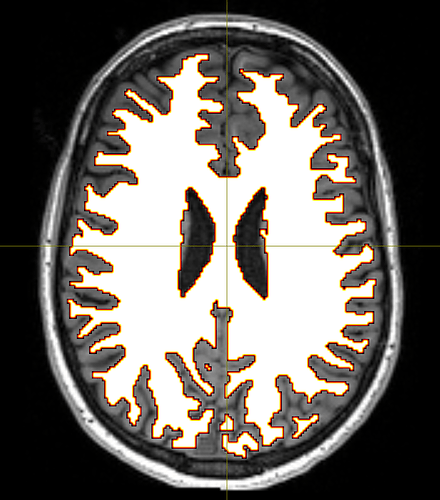
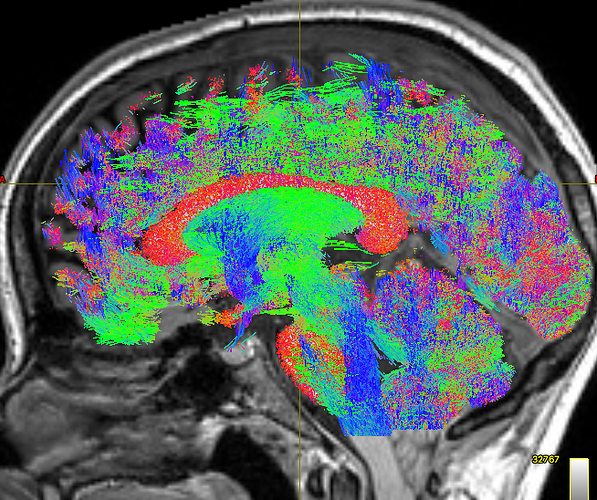
As you can see, the body of the corpus callosum lacks streamlines. I tried using -backtrack and reducing the fod amplitude threshold to 0.05, and got slightly better results:
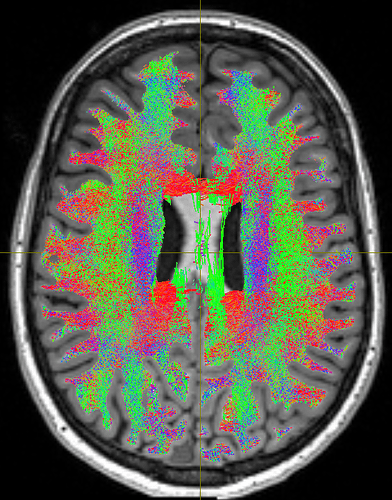
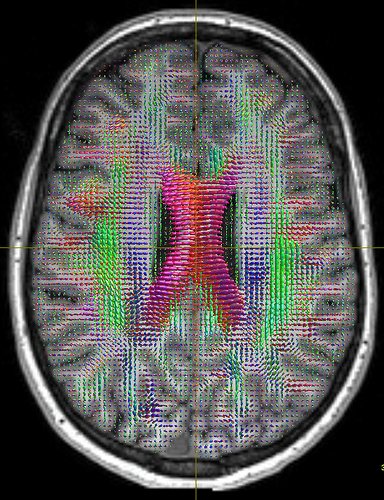
But still, the corpus callosum lacks streamlines. I checked the white matter mask from the 5tt file and tracked streamlines without ACT to make sure the problem is from ACT, and both were fine:
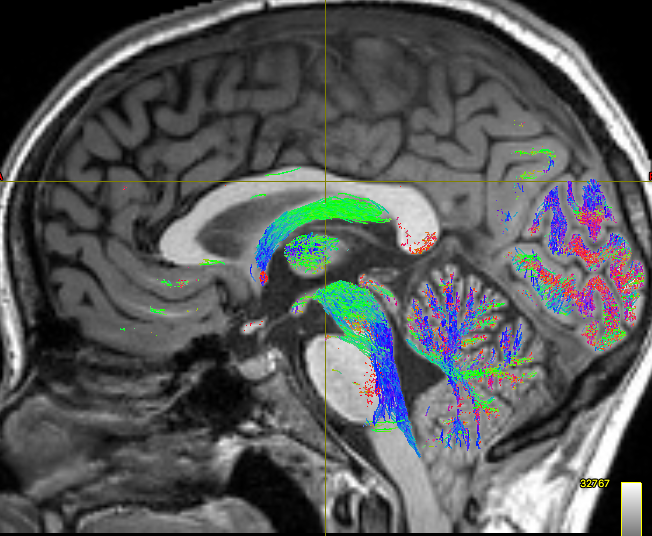
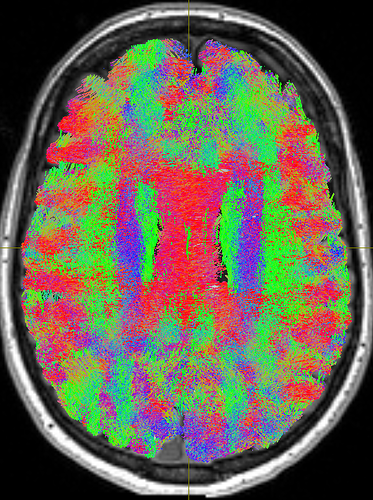
I think the problem is that fibers of the corpus callosum extend into the anatomical CSF compartment due to some volume averaging. Here’s an the fod map to show you what I mean:
Do you have any idea what could be causing this? Is my assumption about the volume averaging accurate?
Looking forward to hearing what everyone has to say. Thanks for the help and for the great software!
Cheers,
Shereif