Hello all,
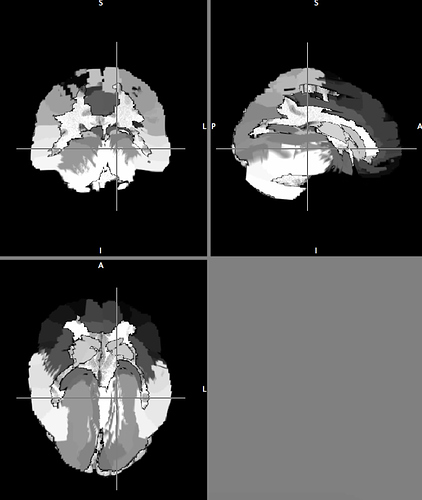
I want to use the HCP data to construct connectome of AAL atlas, I have registered subject’s T1 to DWI, then using flirt and fnirt to register the AAL atlas in MNI space to subject’s T1 space to serve in tck2connectome, but the output AAL2T1 was weird, could anyone help me?
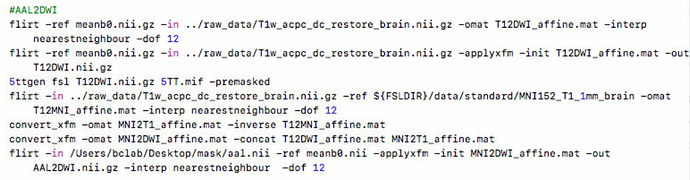
flirt -ref meanb0.nii.gz -in ../raw_data/T1w_acpc_dc_restore_brain.nii.gz -omat T12DWI_affine.mat
flirt -ref ${FSLDIR}/data/standard/MNI152_T1_1mm_brain -in ../raw_data/T1w_acpc_dc_restore_brain.nii.gz -omat T12MNI_affine.mat
convert_xfm -omat MNI2T1_affine.mat -inverse T12MNI_affine.mat
convert_xfm -omat MNI2DWI_affine.mat -concat T12DWI_affine.mat MNI2T1_affine.mat
fnirt --ref=../raw_data/T1w_acpc_dc_restore_brain.nii.gz --aff=MNI2DWI_affine.mat --in=${FSLDIR}/data/standard/MNI152_T1_1mm_brain --cout=MNI2DWI_nonliear
applywarp --ref=meanb0.nii --in=/Users/bclab/Desktop/mask/aal.nii --warp=MNI2DWI_nonliear --out=aal2DWI.nii.gz