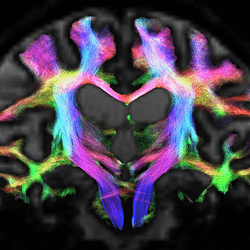
Thanks for the tips. I have made some changes to the workflow to get from the DWI and Freesurfer-T1 to an ACT-informed connectome matrix.
I have pasted the steps and outputs for a single subject with a neurodegenerative disease. Will appreciate any comments especially with regards to the tckgen step. The next thing I will like to do is to weight these matrices by (a) mean FA and to (b) normalise the streamline counts by the average volumes between 2 regions 
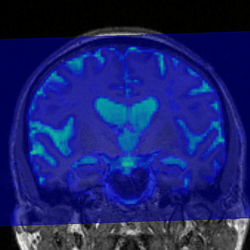
A. Registration of T1 and aparc+aseg.mgz to processed DTI
-
recon-allthe T1 data with Freesurfer -
bbregisterb0 images to orig.mgz / brainmask.mgz -
tkregisterto check/edit registration. This matrix will be used to perform inverse registration of the T1 to DTI space without losing resolution. -
Refer to another MRtrix thread for structural-diffusion registration
Blue = DWI, Greyscale = Aligned T1
B. DWI processing
-
Denoise DWI
dwidenoise i.mif i_denoised.mif;
-
Process diffusion data and perform bias-correction
dwipreproc -0 i_denoised.nii i_preproc.mif -rpe_none
dwibiascorrect i_preproc.mif i_biascorrected.mif -fsl
-
Estimate response function
dwi2response tournier i_biascorrected.mif i_response.txt

-
FOD
dwi2fod csd i_biascorrected.mif i_response.txt i_fod.mif
C. T1 Processing (after Freesurfer and Registration to DWI)
-
Derive tissue-segmented image
5ttgen freesurfer i_aparc+aseg_mrtransformed.mgz i_freesurfer_5TT.mif -
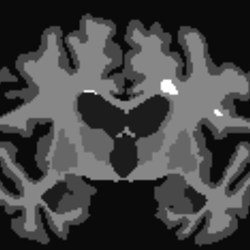
Visualise 5TT
5tt2vis i_freesurfer_5TT.mif i_freesurfer_5TT_vis.mif
@rsmith: Does this look right? The white spots (intensity = 2) correspond to locations of lesions on the WM. How can I deal with them?
I have also tried 5ttgen -fsl but the I am struggling to get good BETs for the 5TT with over-invasive cuts into the cortical GM… -
Modify the integer values in the parcellated image so that the numbers in the image corresponds to the rows and cols of the connectome
labelconvert i_aparc+aseg_mrtransformed.mgz /Applications/freesurfer/FreeSurferColorLUT.txt /Users/MacPro/mrtrix3/src/connectome/tables/fs_default.txt i_nodes.mif
C. Connectome generation
-
Streamline tractography
tckgen i_fod.mif i.tck -act i_FS_5TT.mif -backtrack -crop_at_gmwmi -seed_dynamic i_fod.mif -number 1M -maxlength 250 -
Perform SIFT
tcksift i.tck i_fod.mif i_sift.tck -act i_FS_5TT.mif -term_number 500000
-
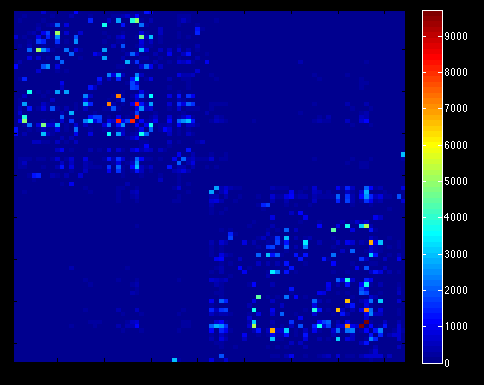
Map the streamlines to the parcellated T1 to produce the connectivity matrix
tck2connectome i.tck i_nodes.mif i_connectome.csv -zero_diagonal
Transform the matrix into a symmetric form where M is your matrix
M = M + triu(M,1)'<-- Thanks, @rsmith ! [see MRtrix thread]
-
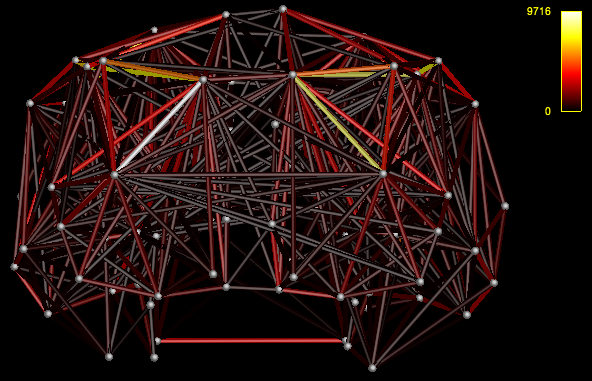
View the connectome
mrview i_nodes.mif -connectome.init i_nodes.mif -connectome.load i_connectome.csv
-
Load connectivity matrices in MATLAB for analyses with Brain Connectivity Toolbox