Hi everyone I’m doing dwi2response command… but I got strange error message like this
could you please tell me how can I solve it?
Hi everyone I’m doing dwi2response command… but I got strange error message like this
could you please tell me how can I solve it?
Hi @CarlisleYun79,
Thanks for your interest in and usage of dwi2response dhollander. The error itself suggests there would be too few b-values in your data. Note that you only need 2 b-values though, and this includes the b=0 data which you would typically always have. But this seems to be the bit where it stumbles: just before that red error line, it’s telling us that you would only be having b=500 data (26 volumes or gradient directions; is that correct?), but no b=0 data. That’s quite unusual indeed.
The bit where I’m worried it might go wrong (just slightly guessing though) would be that you provide your gradient directions and b-values still manually via -fslgrad at this point in your pipeline. I’m not 100% sure this is causing the issue here, but this can in any case be avoided though (and who knows, this might solve the issue here). It might even be that the bet_eddy_correct_info.mif file you’ve got there, already potentially has the gradient information in it, depending on how you preprocessed the data. Let’s first make sure we figure out what’s in that file (and what’s not). Can you run the following commands, and for each of them separately report what output you get on your screen?
First, trying mrinfo directly on the file:
mrinfo bet_eddy_correct_info.mif
Then, checking whether there’s a gradient table there and looking at what it is in full:
mrinfo bet_eddy_correct_info.mif -dwgrad
Then the same thing with your provided bvecs and bvals:
mrinfo bet_eddy_correct_info.mif -fslgrad sub-OAS30004_ses-d1101_dwi.bvec sub-OAS30004_ses-d1101_dwi.bval
And finally seeing how those might be interpreted together with the data file:
mrinfo bet_eddy_correct_info.mif -fslgrad sub-OAS30004_ses-d1101_dwi.bvec sub-OAS30004_ses-d1101_dwi.bval -dwgrad
In general, you want to convert your data into .mif format as early as you can in your entire processing pipeline, and when doing that (via mrconvert) immediately and only once supply the bvecs and bvals via -fslgrad. Once you’ve done that, you can continue working only with the .mif output from that for any subsequent steps, as long as you use MRtrix commands only. The gradient table (the information that is stored in those bvecs and bvals) is in that initial mrconvert step stored for you within the .mif file itself, together with the image data. In that way, there’s only 1 file you need to handle down the track, and the gradient table sticks closely too it (and also, the gradient table is corrected for you if any steps require changing it). So essentially, doing an early mrconvert in your pipeline to .mif and with -fslgrad, means that you don’t have to worry about it anymore down the track and chances of making any mistakes are reduced / kept to a minimum. Does that make sense? 
In any case, let us know the individual outputs of the above 4 commands: it might give some extra insight into what’s exactly going on with your specific data at hand!
Cheers,
Thijs
Thank you for your answer!!
First, mrinfo bet_eddy_correct_info.mif
also I checked ‘mrinfo bet_eddy_correct_info.mif -dwgrad’
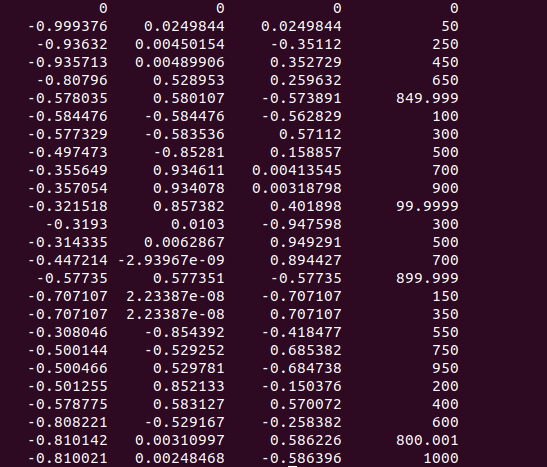
and mrinfo bet_eddy_correct_info.mif -fslgrad sub-OAS30004_ses-d1101_dwi.bvec sub-OAS30004_ses-d1101_dwi.bval is same which I provided…
but I think multi shell is the problem… Am I right…?
then do I have to use dwi2response msmt_5tt?
Well, not really; it’s a bit the opposite (in a way): you data is not “shelled” at all. “Shelled” data only has a very limited number of unique b-values, and lots of gradient directions for each such b-value (and each non-zero b-value is referred to as a “shell” in the literature). In the gradient table, you can see you have a whole range of b-values, and only one or a few directions per b-value.
No, that’s also for shelled data (and as was shown in the past, generally performs less well than dwi2response dhollander).
So in your case, sadly the conclusion is you can’t perform any CSD technique on these data.
Where did you obtain these data from? It’s a very strange dMRI acquisition, I’d say: not many generic things you can do with this. If this is data you’re acquiring yourself (or is being acquired at your institute/…), you should definitely look into this.