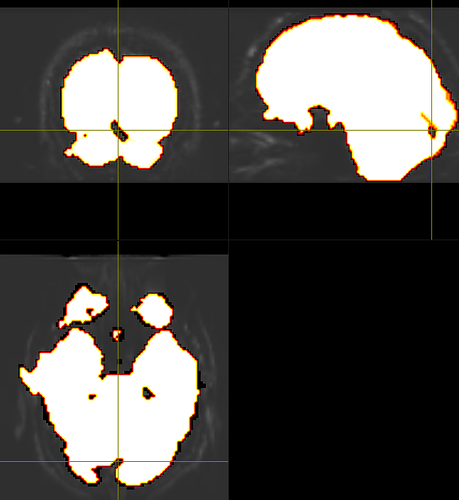
Yes, that mask from dwi2mask algorithm was just from a first pass, and so iterating it between dwibiascorrect and dwi2mask tends to fill in most of the holes (as suggested here). Though, with my dataset, I still tend to have small holes throughout the brain mask, usually between the occipital lobe and cerebellum, even after the second or more mask pass, like so:
This seems to also be the case after extracting a mask from the upsampled the dwi data as well. So, I opted to use FSL’s BET as the main brain mask, while using the mask from dwi2mask for bias field correction for now. Though I do like dwi2mask’s more constrained inclusion of the brain matter comparably, as BET seems to include the meninges too…one of my colleagues is using the ‘erode’ option from mrcalc to constrain the BET mask more, so I might try that out, too.
Just a follow-up question to this - as I am using the mask from dwi2mask primarily for dwibiascorrect, would it be better to use a second bias corrected image from the generation from the second mask? The second mask tends to look more ‘wholesome’ like the one above, but when comparing the bias corrected images, they don’t seem to reveal too many differences…
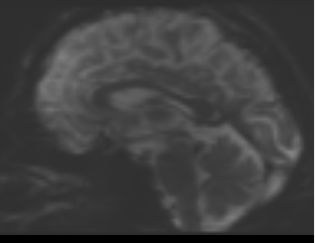 bias corrected image with first pass mask
bias corrected image with first pass mask
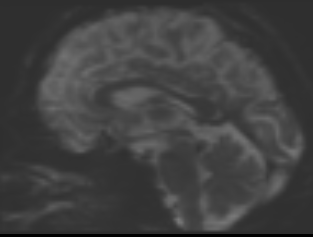 bias corrected image with second pass mask
bias corrected image with second pass mask
Ah, I did not realise that I do not need to manually input a mask for dwibiascorrect…saves me a step! But yes, the mask internally generated within dwibiascorrect seems to be the same as when using dwi2mask.