Dear MRtrix experts,
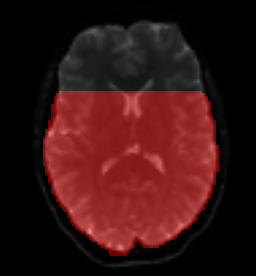
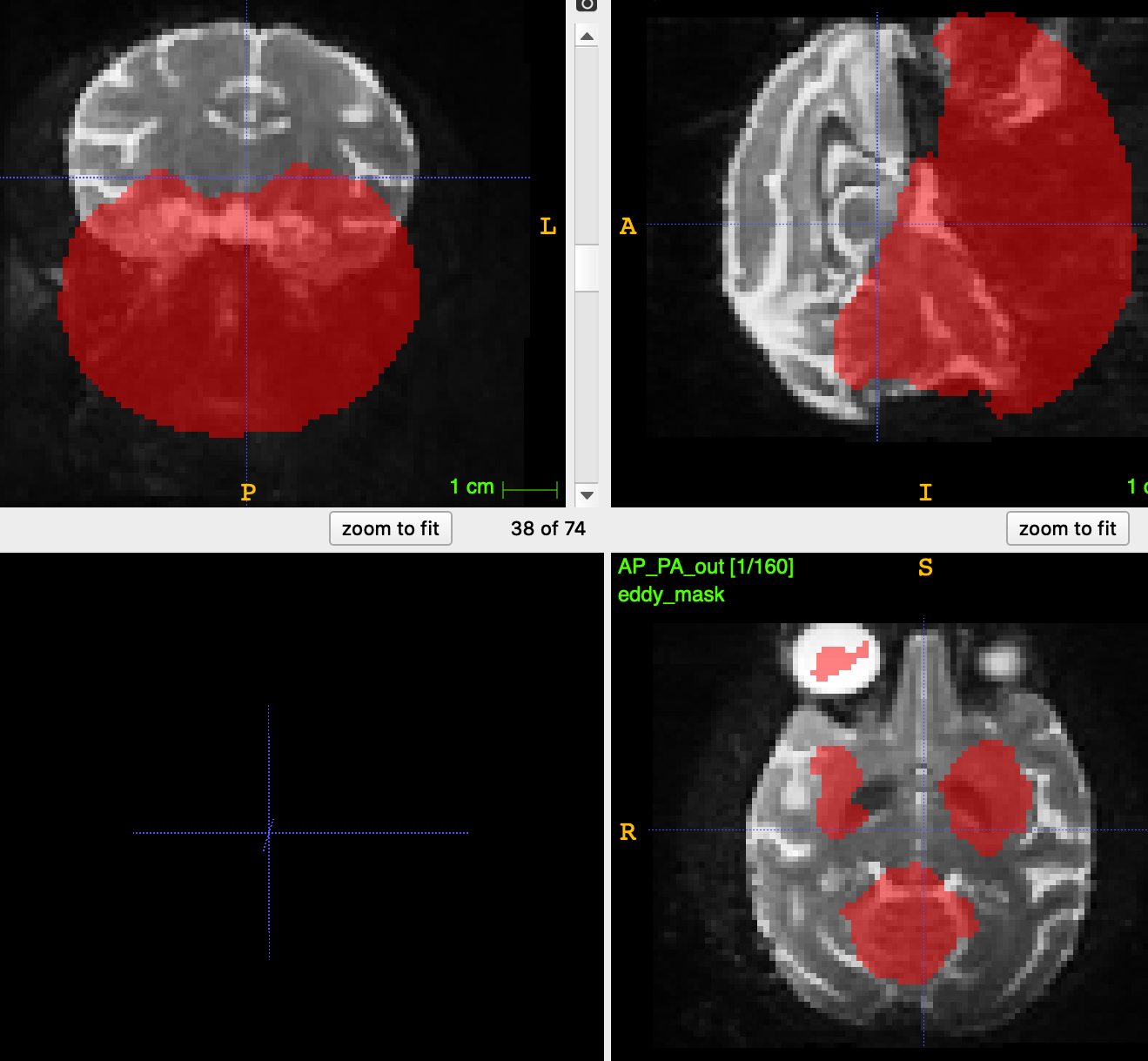
I am currently analysing multishell data (b=0, b=700, b=1200 and b=2800) and I just ran dwifslpreproc (3.0_RC4) on our HPC cluster (the people from HPC installed this version, I don’t really get how, but apparently, this is the newest of newest?). I wanted to check data quality of the output in the eddy output folder and the first thing I opened was the eddy_mask.nii that is created during the process. It looks like a whole part of the brain is cut off, but I have no idea why. As this mask is being used during eddy, I guess this is not good news for the analysis?
This is what the mask looks like (and it is like this for every subject):
For the analysis, I used the following command
dwifslpreproc ${input}/dwi_degibbsed.mif ${input}/geomcorr.mif -pe_dir AP -rpe_pair -se_epi ${input}/b0_revpe.mif -eddy_options " --repol " -eddyqc_all ${input}/eddy/
The information that was saved, is the following:
dwifslpreproc:
dwifslpreproc: Note that this script makes use of commands / algorithms that have relevant articles for citation; INCLUDING FROM EXTERNAL SOFTWARE PACKAGES. Please consult the help page (-help option) for more information.
dwifslpreproc:
dwifslpreproc: Generated scratch directory: /vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/
Command: mrconvert /user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/dwi_degibbsed.mif /vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/dwi.mif -json_export /vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/dwi.json
Command: mrconvert /user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/b0_revpe.mif /vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/se_epi.mif
dwifslpreproc: Changing to scratch directory (/vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/)
Command: dirstat dwi.mif -output asym
Command: mrinfo dwi.mif -export_grad_mrtrix grad.b
dwifslpreproc: 1 spatial axis of DWIs has non-even size; this will be automatically padded for compatibility with topup, and the extra slice erased afterwards
Command: mrconvert se_epi.mif -coord 2 74 - | mrcat se_epi.mif - se_epi_pad2.mif -axis 2
Command: mrconvert dwi.mif -coord 2 74 - | mrcat dwi.mif - dwi_pad2.mif -axis 2
Command: mrconvert se_epi_pad2.mif topup_in.nii -strides -1,+2,+3,+4 -export_pe_table topup_datain.txt
Command: topup --imain=topup_in.nii --datain=topup_datain.txt --out=field --fout=field_map.nii.gz --config=/apps/gent/CO7/skylake-ib/software/FSL/6.0.3-foss-2019b-Python-3.7.4/fsl/etc/flirtsch/b02b0.cnf --verbose
Command: mrinfo dwi_pad2.mif -export_pe_eddy applytopup_config.txt applytopup_indices.txt
Command: dwiextract dwi_pad2.mif -pe 0.0,-1.0,0.0,0.0336 - | mrconvert - dwi_pad2_pe_1.nii -json_export dwi_pad2_pe_1.json
Command: applytopup --imain=dwi_pad2_pe_1.nii --datain=applytopup_config.txt --inindex=1 --topup=field --out=dwi_pad2_pe_1_applytopup.nii --method=jac
Command: mrconvert dwi_pad2_pe_1_applytopup.nii.gz dwi_pad2_pe_1_applytopup.mif -json_import dwi_pad2_pe_1.json
Command: dwi2mask dwi_pad2_pe_1_applytopup.mif - | maskfilter - dilate - | mrconvert - eddy_mask.nii -datatype float32 -strides -1,+2,+3
Command: mrconvert dwi_pad2.mif eddy_in.nii -strides -1,+2,+3,+4 -export_grad_fsl bvecs bvals -export_pe_eddy eddy_config.txt eddy_indices.txt
Command: eddy_openmp --imain=eddy_in.nii --mask=eddy_mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --repol --out=dwi_post_eddy --verbose
dwifslpreproc: Command 'eddy_quad' not found in PATH; not running automated eddy qc tool QUAD
Command: mrconvert dwi_post_eddy.nii.gz result.mif -coord 2 0:74 -strides -2,-3,4,1 -fslgrad dwi_post_eddy.eddy_rotated_bvecs bvals
Function: os.makedirs('/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy')
Function: shutil.copy('dwi_post_eddy.eddy_parameters', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_parameters')
Function: shutil.copy('dwi_post_eddy.eddy_movement_rms', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_movement_rms')
Function: shutil.copy('dwi_post_eddy.eddy_restricted_movement_rms', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_restricted_movement_rms')
Function: shutil.copy('dwi_post_eddy.eddy_post_eddy_shell_alignment_parameters', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_post_eddy_shell_alignment_parameters')
Function: shutil.copy('dwi_post_eddy.eddy_post_eddy_shell_PE_translation_parameters', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_post_eddy_shell_PE_translation_parameters')
Function: shutil.copy('dwi_post_eddy.eddy_outlier_report', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_outlier_report')
Function: shutil.copy('dwi_post_eddy.eddy_outlier_map', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_outlier_map')
Function: shutil.copy('dwi_post_eddy.eddy_outlier_n_stdev_map', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_outlier_n_stdev_map')
Function: shutil.copy('dwi_post_eddy.eddy_outlier_n_sqr_stdev_map', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_outlier_n_sqr_stdev_map')
Function: shutil.copy('dwi_post_eddy.eddy_outlier_free_data.nii.gz', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_outlier_free_data.nii.gz')
Function: shutil.copy('eddy_mask.nii', '/user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/eddy/eddy_mask.nii')
Command: mrconvert result.mif /user/gent/412/vsc41214/scratch_vo_user/LAT_study/subjects/LAT_072/geomcorr.mif
dwifslpreproc: Changing back to original directory (/vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study)
dwifslpreproc: Deleting scratch directory (/vscmnt/gent_kyukon_scratch/_kyukon_scratch_gent/vo/000/gvo00047/vsc41214/LAT_study/dwifslpreproc-tmp-ZNUOJV/)
To me, the output did not look weird.
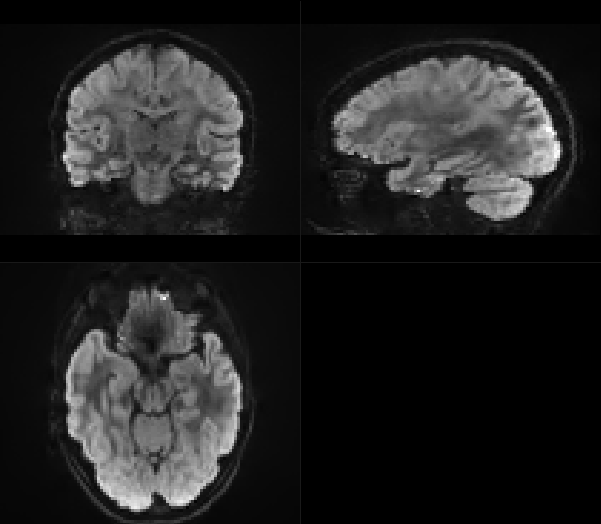
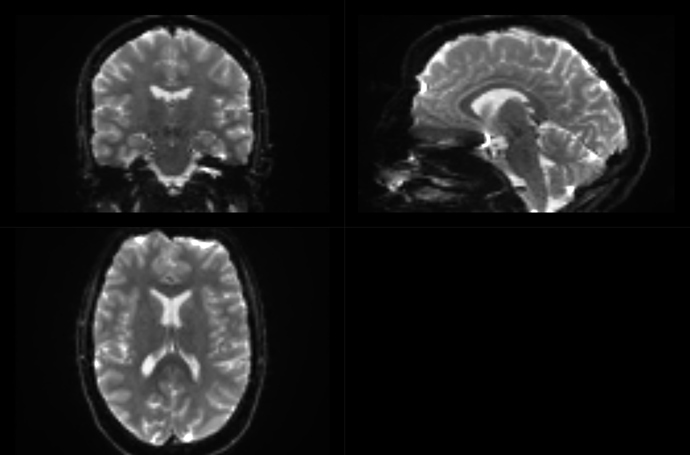
This is before dwifslpreproc:
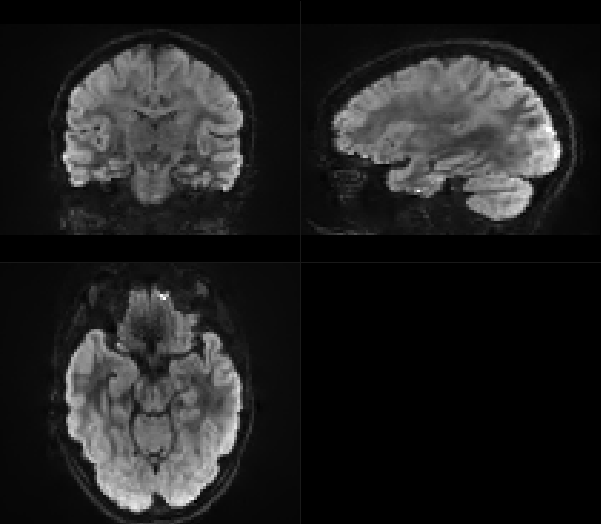
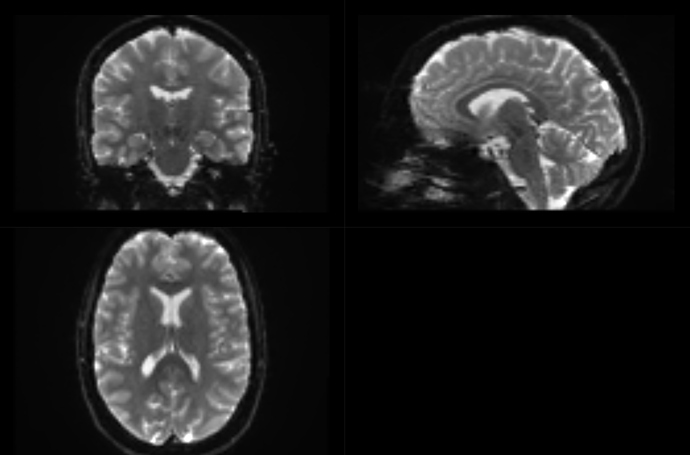
And after dwifslpreproc:
Did something go wrong? And if so: what went wrong and how can I resolve this?
Thank you in advance for checking my post!
Best,
Helena
 I will send it to your email addresses. Unfortunately I am not able to re-upload or send it via another way at the moment as I am traveling right now. Will be back behind a computer on the 28th.
I will send it to your email addresses. Unfortunately I am not able to re-upload or send it via another way at the moment as I am traveling right now. Will be back behind a computer on the 28th.