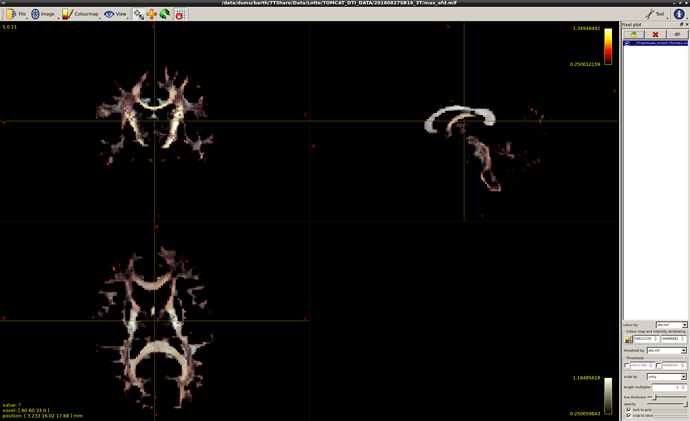
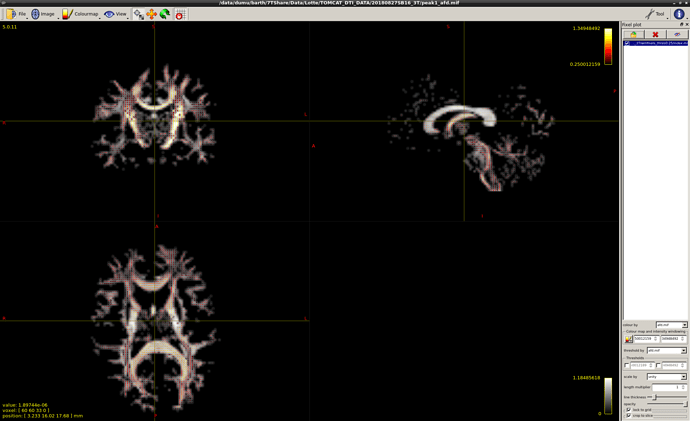
Ok, just double-checked myself to be certain: you’re right, they’re not at all times guaranteed to be sorted from large to small (I wasn’t aware of that myself when I typed up any of the above!). Other than that though, the extracted values do match the AFD values from the original fixels. But basically, to get what you’re after in the example you provide, you can’t rely on the ordering in which they are stored.
Looking further into the options, I was then (now) thinking you could get this nonetheless by adding the -weighted option to fixel2voxel, and providing that option with the same afd.mif file. The idea here would be that the value you extract (the main argument to the command) is separate from the data that defines the “size” and that the latter would be used for sorting. However, trying that myself, I encountered a warning:
fixel2voxel: [WARNING] Option -weighted has no meaningful interpretation for the operation specified; ignoring
This is strange as there’s also the -number option to limit the extraction to a given (maximum) number of fixels. The documentation of this option reads:
-number N
use only the largest N fixels in calculation of the voxel-wise statistic;
in the case of "split_data" and "split_dir", output only the largest N
fixels, padding where necessary.
Suggesting at least a notion of sorting (otherwise there’d be no notion of the “largest N fixels”). But while that option does limit the number of extracted fixels, it still doesn’t sort them; probably also implying that its promise of extracting the largest N fixels might be broken (though I haven’t tested that). Finally, I tried to combine -number and -weighted, so as to explicitly introduce the notion of size again, hoping that -number would use it. But alas, the same warning popped up, and the output didn’t sort; so I’m again also left wondering whether the largest N promise holds here…
This is hard to check because most of the time the fixels do come out “almost” sorted. I suppose they just come out in the order they’re stored in the fixel data. That fixel data itself in turn probably gets its ordering just from how the fixels are segmented, but because the algorithm has a few rules about merging or separating individual peaks into single fixels based on relative amplitudes of peaks and valleys, I can imagine the final fixels don’t come out per se in the order the original separate peaks were encountered (e.g. when peaks are merged, I suppose). Lots of supposing and guessing here on my part in this little paragraph… so don’t take it for granted (but it does seem sensible to me in this way). @rsmith can probably help you out with the exact behaviour or hidden/unexpected quirks of fod2fixel followed by fixel2voxel.
However, that aside, the thing I did encounter with the -number not sorting makes me strongly wonder/question whether the “largest N fixels” promise there actually holds in practice, especially when the -weighted is sticking to the warning of not being meaningful for split_data… Even if this does still check out; the documentation could certainly be improved a bit, e.g. to at least mention when sorting can (not) be expected or relied upon. I’ll create another issue to sort out the documentation, or bug if there is one.
Cheers,
Thijs