Hi all,
I am currently trying to work on HCP 7T diffusion MRI data but happen to find out that the b0 signal in the frontal and temporal lobes are very small (those areas appear to be very dark). This causes serious problem for the next step of model fitting in those areas. For example, the FA map looks wired. Is there any way to tackle with this problem?
Thanks a lot in advance~~~
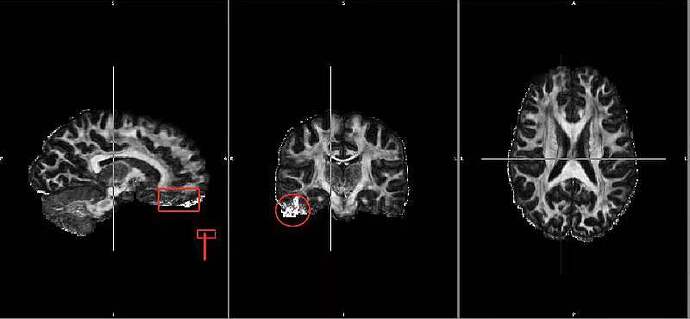
Here is an exemplary FA map from HCP 7T diffusion dataset and the areas in the red rectangle shape are those problematic areas I meant…
I think the areas you’re highlighting are where you’d expect strong susceptibility-induced distortions, due to field inhomogeneities. These will always be problematic areas to image, especially at high resolution. The strategy for the HCP was to acquire the data twice with reversed phase-encoding direction, running left-right / right-left (which is unusual, most people use anterior-posterior), and use FSL’s topup & eddy tools to try to correct for these distortions – but as you can clearly see, in some regions it’s not possible to recover the signal accurately. I don’t think there’s very much that can be done here, unfortunately…
1 Like
Could also be some signal drop off in temporal regions due to B1 transmit inhomogeneity. but yes as Donald says you can’t do much. You could try doing your own pre-processing with eddy and topup if you can get hold of the unprocessed data.
1 Like
Thanks a lot for your reply! Yes, it is partly due to the susceptibility induced distortions. How is the St Thomas’ 7T working? Does it give similar things and if so, is anyone working on improving it? I heard from Siemens scientists that ptx might help. Do you guys use it when you acquire 7T diffusion?
Thanks a lot for your reply. yea, a bit sad to know that there is not much I can do with this HCP 7T diffusion dataset. I don’t think I can come up with a better solution with this problem than Topup and Eddy from the mri physical perspective though, as it is not my expertise. Hopefully some MRI physicists might fix this problem in future soon as 7T now is taking off in research and clinical practice…
I have to admit I’ve not starting using the 7T system yet – but there’s plenty of other very capable MR physicists working on it.
To answer your question: if the issue is susceptibility-induced distortion, I don’t think pTx is going to help at all – it won’t compensate for B0 inhomogeneities. But if there are B1 field inhomogeneities as Brad suggests, then yes, pTx should help with that, and may recover signal that would otherwise be lost.
Ah I see. Good to know there are MRI physicists working on it. Hopefully this can be solved soon. Thanks for your reply again~