Hi everyone,
I want to reconstruct tracts within a specific cortical region, ie. the seed region and endpoint region are both the specific cortical region.
I have tried tckgen wmfod.mif tracks_1.tck -act 5tt_coreg.mif -backtrack -select 100000 -seed_image mask -include_ordered mask -seed_unidirectional
and double “-include_ordered”:
tckgen wmfod.mif tracks_1.tck -act 5tt_coreg.mif -backtrack -select 100000 -seed_image mask -include_ordered mask include_ordered mask -seed_unidirectional
But none of the above get the good result 
So what should I do?
Thanks in advance for any advice,
Panshi
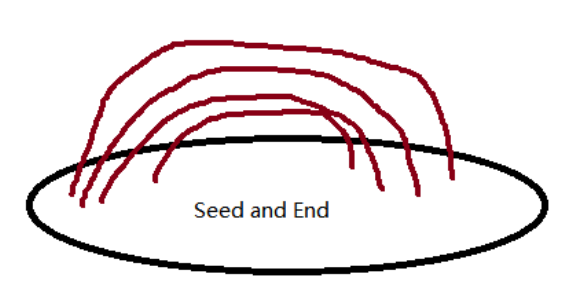
Supplementary diagram: I want to reconstruct the brown tracts
Hi Panshi
What qualifies as a good / bad result?
I think having identical masks for seeding and including is not going to get you anywhere, since any streamline will have already fulfilled your inclusion criteria simply by existing…
I would suggest using two ROIs (you can make them very thin so in anatomical terms they’re practically identical), with the include one “behind” the seed one, and using the -stop flag. Maybe also use -maxlength to get only “local” streamlines.
e.g. I used the following command:
tckgen fod.mif -seed_image yellow_roi.mif -include pink_roi.mif -seed_unidirectional -select 100 -stop -maxlength 50 test_out.tck
to get this (crosssection view; ROIs are single slice disks):
![]()

