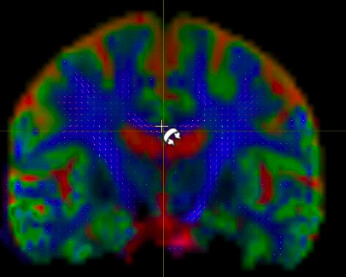
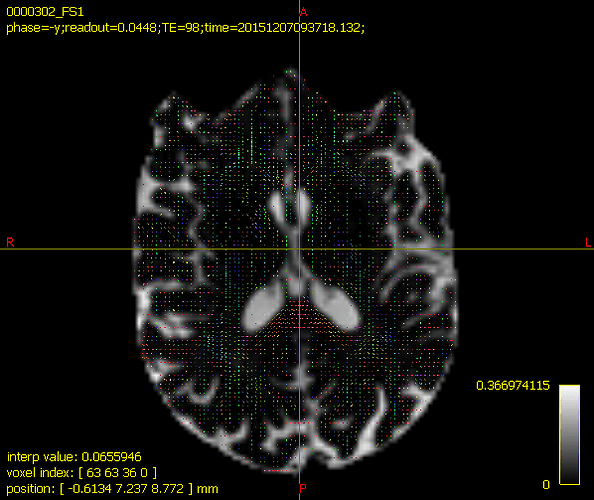
I have dwi data with b=0,1000 and following the BATMAN tutorial, I noticed the FODs seem correct but the color coding of tissue types is missing.
My commands:
dwi2response dhollander ${dir_out_sub}/dwi_${subject}_preproc_unbias.mif response_wm_${subject}.txt response_gm_${subject}.txt response_csf_${subject}.txt -voxels voxels_${subject}.mif -force
dwi2fod msmt_csd ${dir_out_sub}/dwi_${subject}_preproc_unbias.mif response_wm_${subject}.txt wmfod_${subject}.mif response_csf_${subject}.txt csffod_${subject}.mif -mask ${dir_out_sub}/dwi_${subject}_mask.mif -force
mrconvert -coord 2 0 wmfod_${subject}.mif -| mrcat csffod_${subject}.mif - vf_${subject}.mif -force
Then to view I write:
ds: mrview vf_${subject}.mif -odf.load_sh wmfod_${subject}.mif
Is there something I am missing or is it just not possible to view the tissue color segmentation like so: