In my attempts to re-scale b-values for a multi-shell diffusion series of mine (using a pipeline that forces me to scale b-values), I am encountering differences in the bvecs exported by MRtrix3 and the original bvecs that were imported.
For background, I am working with the ABCD GE RSI sequence, which has some peculiarities in how it stores diffusion bvecs in the GE dicom header. That is, from a user’s perspective, when converting with dcm2niix, only the largest bvalue will be extracted (i.e. if your sequence has bvals of 500, 1000, and 4000, bvals created by dcm2niix will only have 4000). To correct this, bvecs need to be read from an external tensor.dat file, provided by GE, which one can then derive the “correct” bvals from.
After running the procedure above on our diffusion data, the image was fed into the previously mentioned pipeline. In attempting to undo the bvalue-scaling of the preprocessed output, I figured I could just re-derive the bvals from the bvecs as before:
bvals = float(max_bval) * np.sum(np.power(bvecs, 2), axis=1)
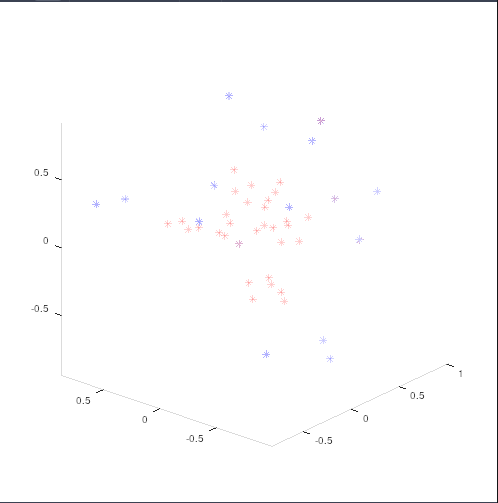
Doing so, yielded only bvals of 4000, which does not make sense. After testing with unprocessed output, lo and behold, exported gradient tables from freshly mrconvert’ed files are different from the “imported” gradient tables. This is true even if I turn off bvalue-scaling.
(imported in red, exported in blue)
I was curious, how is MRtrix3 scaling imported bvec tables?
(tests performed with latest git version of mrconvert – 3.0.4-10-gf633dfd7. Data originally processed with MRtrix3 version 3.0.1)