Dear MRtrix3 team,
I’ve been working through the HCP connectome tutorial, and I’ve come across this odd behaviour.
When I run:
dwi2response msmt_5tt DWI.mif 5TT.mif RF_WM.txt RF_GM.txt RF_CSF.txt -voxels RF_voxels.mif
I get response function outputs that do not match the order in which they are supposed to come out. This page tells me the output order is WM, GM, CSF but when I inspect RF_voxels.mif, the voxels correspond as follows:
- Volume 0 = CSF
- Volume 1 = cortical GM
- Volume 2 = WM
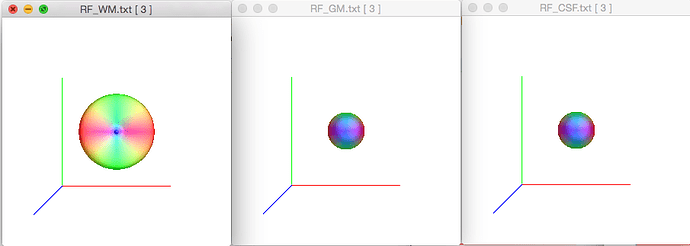
This also bears out in response function files when I inspect them with shview, screenshot here (note the title of each window).
I checked the first obvious culprit (5TT.mif) but it looks fine, with the correct tissue order as described here (cortical GM, subcortical GM, WM, CSF, pathological tissue). I also made a fresh git pull to ensure everything is up to date.
Any ideas as to what might be going on here?
Hi @IvanAlvarez,
There’s nothing wrong here; I’ll explain what you’re seeing!
-
The output order of the response function files is certainly WM-GM-CSF, but that of the -voxels image is CSF-GM-WM. This is because the -voxels image is meant to be an RGB image, so you can see the voxels in mrview in the same colours as used in e.g. the MSMT-CSD paper that started showing these voxels (and tissue maps) as CSF = red, GM = green, WM = blue. Several other publications thereafter have adopted the same colour “convention”.
-
Your response functions should be fine, but due to a bug in shview, the GM and CSF response functions (or any isotropic multi-shell response really) won’t show correctly: it misinterprets the l=0 coefficients per shell as if there’s only one shell with those coefficients as a vector. Long story short: (currently) don’t use shview to look at your GM and CSF responses. Even if it would work correctly, each shell would only show as a sphere, which is not that informative anyway. Checking GM and CSF responses is better done by directly opening the file in a text editor, or even plotting the numbers in there using, e.g., Matlab or Excel. Your WM response function is actually showing correctly, but you’re only seeing the first (b=0) shell there, which is indeed just a sphere. You can use the right and left arrow keys to navigate to the other shells. Hitting right arrow a couple of times, you should see disc shapes with increasing “sharpness”.
-
We’ve recently introduced a new algorithm in dwi2response that should be much less hassle to use, and often gives you more accurate results, especially for the CSF response. It doesn’t rely on a co-registered T1w image and 5TT segmentation thereof. To use it (with your file names), you’d just do this:
dwi2response dhollander DWI.mif RF_WM.txt RF_GM.txt RF_CSF.txt -voxels RF_voxels.mif
I can confirm it increases accuracy of the CSF response significantly for the preprocessed HCP data.
Hope this helps!
Cheers,
Thijs
@Thijs: guess it’s time we tackled this issue, then?
Thanks @Thijs, that explains a lot!
Ah right, it’s the colour convention that threw me off. As you say, I went back and my data does match the MSMT-CSD paper and shview for the WM response function does display as you described once I hit left/right arrows. May I suggest documenting that functionality in the shview help page? 
I just ran dwi2response dhollander as you suggest and I get pretty good results which is encouraging! Thanks for the suggestion, will play around with it a bit more and see how far I get.
The shview bugs should be gone now!
@IvanAlvarez: if you update your MRtrix3 installation (git pull and ./build), shview should no longer show those weird shapes upon opening your GM and CSF response functions. About the left/right arrows functionality: I agree it’s a bit hidden (I only discovered it not so long ago myself!). If you open the File menu in shview though, you’ll see 2 entries to go to the previous and next shells, as well as their keys. As such, the menus of shview and mrview are a bit a manual as well. 
About dwi2response dhollander: yep, we get good performance here to on all HCP data, as well as basically all our other data so far. Maybe we should consider updating those tutorials: there’s currently much more scenarios where dwi2response msmt_5tt will give suboptimal or even quite wrong results; not to mention dwi2response dhollander is faster and easier to use, and relies on much less preprocessing.
@Thijs ok, just did a fresh git pull and ./build and happy to report the weird lobes in GM and CSF response are indeed gone.
I think that’s a good idea, it will save time in the long run answering questions from people like me 
1 Like