Thank you for your patient reply.
Fig 8 is a really cool result! I love it very much.
Do you mean my problem may not only list on the mask, but also FOD?
My bval file is as follows:
In my case, it seems a single-shell DWI, As shown here, ss3t_csd is not available yet. do you mean that I can use "msmt_csd" instead of "csd" when I use dwi2fod to fiber orientation distribution estimation? Just like the document, so that I can strike out the gray matter.
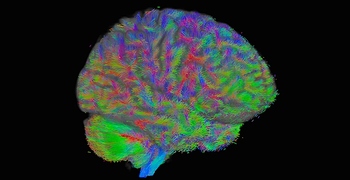
By the way, Figure 9 is really fantastic. However, Global Tractography seems to run on a cohort of images. I just test one single DWI data, does Global Tractography fit me? I want to try it then. 
Fig 9: (Global Tractography)
Many Thanks,
Chaoqing