Hi, @NicDC
Many thank you for your reply. ![]()
Follow your suggestion
I have upgraded my code as followed, and it works perfectly!!
flirt -ref ../MNI152_T1_1mm_brain.nii.gz -in I0image.nii.gz -omat template2mni.mat -dof 12
fnirt --ref=../MNI152_T1_1mm_brain.nii.gz --in=I0image.nii.gz --aff=template2mni.mat --cout=warps_template2mni
invwarp --ref=I0image.nii.gz --warp=warps_template2mni --out=warps_mni2template
warpinit ../MNI152_T1_1mm_brain.nii.gz inv_identity_warp_no_mni.nii
applywarp --ref=I0image.nii.gz --in=inv_identity_warp_no_mni.nii --warp=warps_mni2template.nii.gz --out=mrtrix_warp_mni2template.nii.gz
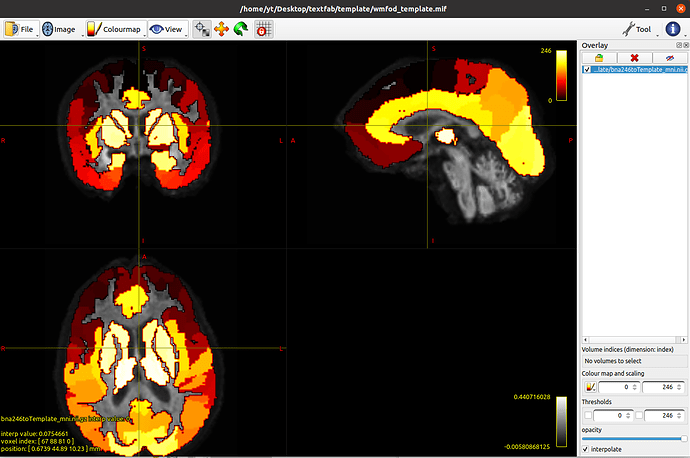
mrtransform ../BN_Atlas_246_1mm.nii.gz -warp mrtrix_warp_mni2template.nii.gz bna246toTemplate_mni.nii.gz -template wmfod_template.mif -interp nearest
mrconvert bna246toTemplate_mni.nii.gz bna246toTemplate_mni.mif
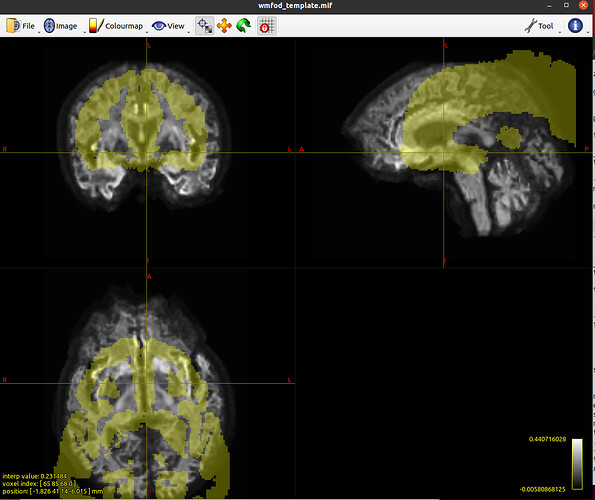
And given the thread you quoted, I’ve tried exactly as what the @maxpietsch suggested, and worked successfully, but the results were even weirder ![]() . After applying the warp to the Atlas, some labels disappeared. I have found that some regions disappeared in the warp_MNI152.mif, but I’m not familiar with ANTs, so I have no idea how to optimize it, would you please guide me?
. After applying the warp to the Atlas, some labels disappeared. I have found that some regions disappeared in the warp_MNI152.mif, but I’m not familiar with ANTs, so I have no idea how to optimize it, would you please guide me?
## warp image use ANTs nonlinear
#transform the template_mask.mif to the ANTs compatible format
mrconvert template_mask.mif template_mask.nii.gz
# register MNI152 to the I0image
antsRegistration --verbose 1 --dimensionality 3 --float 0 --output [ants,antsWarped.nii.gz,antsInverseWarped.nii.gz] --interpolation Linear --use-histogram-matching 1 --winsorize-image-intensities [0.005,0.995] --transform Rigid[0.1] --metric CC[I0image.nii.gz,../MNI152_T1_1mm_brain.nii.gz,1,4,Regular,0.1] --convergence [1000x500x250x100,1e-6,10] --shrink-factors 8x4x2x1 --smoothing-sigmas 3x2x1x0vox --transform Affine[0.1] --metric CC[I0image.nii.gz,../MNI152_T1_1mm_brain.nii.gz,1,4,Regular,0.2] --convergence [1000x500x250x100,1e-6,10] --shrink-factors 8x4x2x1 --smoothing-sigmas 3x2x1x0vox --transform SyN[0.1,3,0] --metric CC[I0image.nii.gz,../MNI152_T1_1mm_brain.nii.gz,1,4] --convergence [100x70x50x20,1e-6,10] --shrink-factors 4x2x2x1 --smoothing-sigmas 2x2x1x0vox -x [template_mask.nii.gz,../MNI152_T1_1mm_brain_mask.nii.gz]
# Generate an identity (deformation field) warp
warpinit ../MNI152_T1_1mm_brain.nii.gz identity_warp[].nii
# Transform this identity warp
for i in {0..2}; do
antsApplyTransforms -d 3 -e 0 -i identity_warp${i}.nii -o mrtrix_warp${i}.nii -r I0image.nii.gz -t ants1Warp.nii.gz -t ants0GenericAffine.mat --default-value 2147483647
done
# Correct the warp
warpcorrect mrtrix_warp[].nii mrtrix_warp_corrected.mif -marker 2147483647
# Warp the image
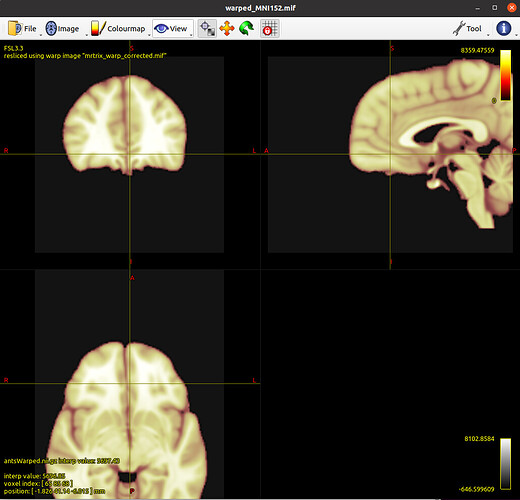
mrtransform ../MNI152_T1_1mm_brain.nii.gz -warp mrtrix_warp_corrected.mif warped_MNI152.mif
# check that the conversion went as expected by comparison with ANTs’ output
mrview warped_MNI152.mif -overlay.load antsWarped.nii.gz -overlay.opacity 0.3
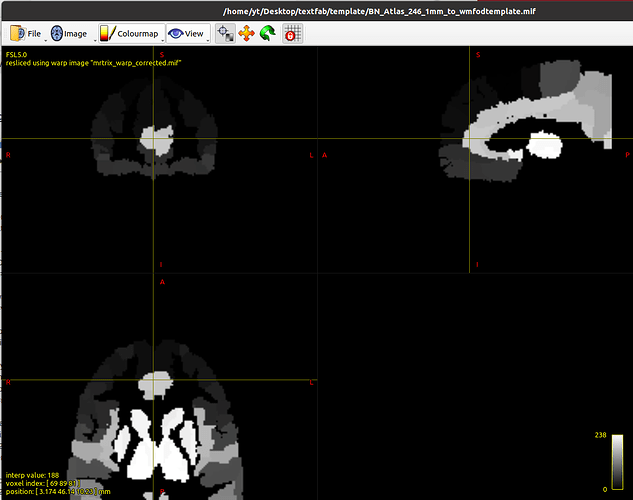
# Apply the warp to Atlas (with regridding)
mrtransform ../BN_Atlas_246_1mm.nii.gz -warp mrtrix_warp_corrected.mif -interp nearest BN_Atlas_246_1mm_to_wmfodtemplate.mif
# Check everything worked
mrview wmfod_template.mif -roi.load BN_Atlas_246_1mm_to_wmfodtemplate.mif -roi.opacity 0.3
Sorry, I got a bit confused. Is there any difference between the registration results reults form the ANTs and the FSL? They seem the same when overlaying the results (i.e. Atlas2wmfod_template.mif) with the wmfod_template, ideally. So I can also using tck2connectome and connectome2tck with the warped results (i.e. Atlas2wmfod_template.mif) which I generated from FSL, right?
Sorry for the lengthy questions, and thank you very much for your patience and kindness.
Best wishes,
Yuting.