Hi, Donald,
1. ACT with data2(An example data)
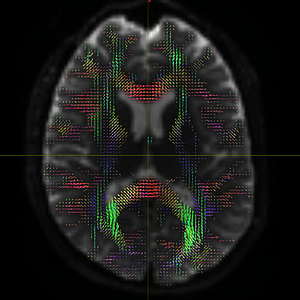
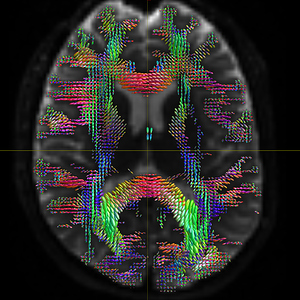
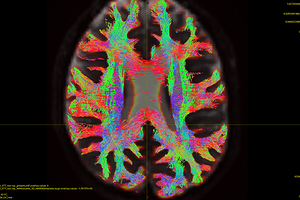
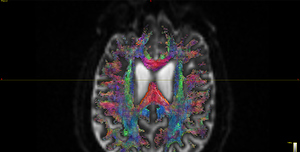
The FODs ( HARDI150_WMvolume_fod.mif ) is not in expected anatomical directions(as shown in Fig. 5.1).
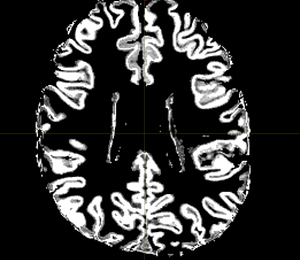
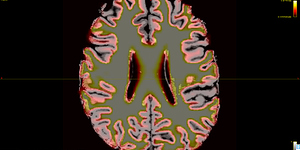
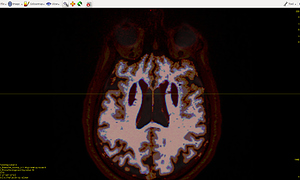
The 5TT image (T1_HARDI150_5TT_nocrop.mif) is shown in Fig.5.2.
However, I cannot fix it by using mrconvert -stride command, I tried:
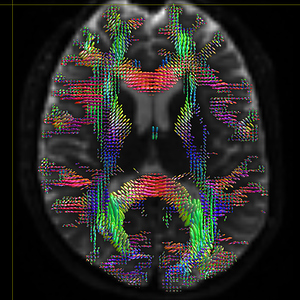
mrconvert HARDI150.nii.gz -stride -1,2,3,4 HARDI150_StrideX.nii.gz ( Fig.5.3 )
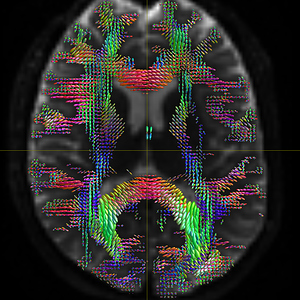
mrconvert HARDI150.nii.gz -stride 1,-2,3,4 HARDI150_StrideY.nii.gz ( Fig.5.4 )
mrconvert HARDI150.nii.gz -stride -1,-2,3,4 HARDI150_StrideXY.nii.gz ( Fig.5.5 )
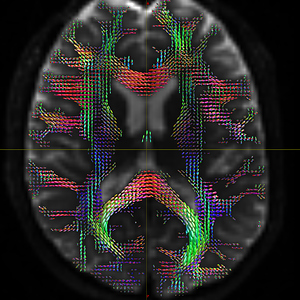
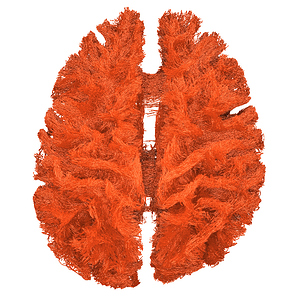
I finally fix HARDI150 FODs by inverse the second column of HARDI150.bvecs as shown in Fig 5.6 ( HARDI150.bvecs maybe not generated by Mrtrix3 ). Fig 5.7 shown the corpus callosum region. However, it is not able to generate the fibers in corpus callosum region( see Fig 5.9 and Fig 5.10 ).
Do you have any ideas? I still cannot find the solution.
2. ACT with data1
The DTI_2013.bvecs is created by Mrtrix3 command:
mrconvert 2013-02-06_10_56_37.0/ -export_grad_fsl DTI_2013.bvecs DTI_2013_bvals DTI_2013.nii.gz
① In native anatomical space:
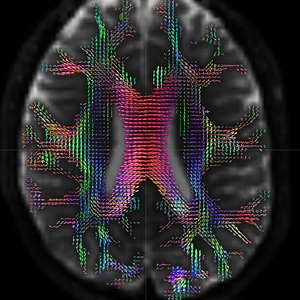
DTI_2013.nii.gz is 4D image and t1_2013.nii.gz is the corresponding 3D image. I use the original data: ( DTI_2013.nii.gz / t1_2013.nii.gz ) to generate the fod image ( DTI_2013_fod.mif ) , the fod image correctly shows the anatomical direction, see Fig 6.1.
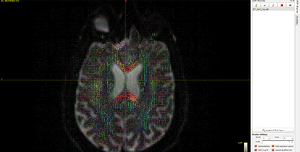
Then I do recon-all command of FreeSurfer to generate cortical parcellations of t1_2013.nii.gz. In this way, the t1_2013.nii.gz data will naturally go to FreeSurface space ( t1_2013_freesurfer.nii.gz ) . So I register DTI_2013.nii.gz to DTI_2013_freesurfer.nii.gz, following Lucius’s instruction, They seems registered nicely, see Fig 6.2:
A. Register DTI_b0 to T1_freesurfer:
flirt -in DTI_2013_b0.nii.gz -ref t1_2013_freesufer_skulled.nii.gz -omat DTI_2013_b0_to_t1_2013_freesurf_skulled.mat
flirt -in DTI_2013_b0.nii.gz -ref t1_2013_freesufer_skulled.nii.gz -applyxfm -init DTI_2013_b0_to_t1_2013_freesurf_skulled.mat -out DTI_2013_b0_freesurfer.nii.gz
B. Register DTI to T1_freesurfer:
flirt -in DTI_2013.nii.gz -ref t1_2013_freesufer_skulled.nii.gz -applyxfm -init DTI_2013_b0_to_t1_2013_freesurf_skulled.mat -out DTI_2013_freesurfer.nii.gz
② In FreeSurfer space:
I use the data ( DTI_2013_freesurfer.nii.gz/ t1_2013_freesurfer.nii.gz ) to generate the fod image (DTI_2013_freesurfer_fod.mif), From then on, the fod image can not correctly indicate the anatomical direction, see Fig 6.3. You can find the tensors on the top half the region of corpus callosum are incorrect. (It seems half of the orientation is correct and the other half goes wrong).
③ In MNI152 space:
Then I use the similar registration command to register (DTI_2013_freesurfer.nii.gz/ t1_2013_freesurfer.nii.gz) to MNI152 space. The fod image ( DTI_2013_freesurfer_in_MNI152_fswm_fod.mif ) certainly incorrectly indicates the anatomical direction, as shown in Fig 1.3.
Does it mean I did wrong registration? I tried mrconvert -stride and inverse the second collum of bvecs, the output fod images maintain wrong. I hope to know how does it happen and expect to figure it out.
Sweet Thanks,
Chaoqing
Fig 5.1 ( HARDI150_WMvolume_fod.mif )
Fig 5.2 ( T1_HARDI150_5TT_nocrop.mif )
Fig 5.3 ( HARDI150_StrideX_WMvolume_fod.mif )
Fig 5.4 ( HARDI150_StrideY_WMvolume_fod.mif )
Fig 5.5 (HARDI150_StrideXY_WMvolume_fod.mif )
Fig 5.6 (HARDI150_bvecInverseY_WMvolume_fod.mif )
Fig 5.7 (HARDI150_bvecInverseY_WMvolume_fod.mif )
Fig 5.8
( T1_HARDI150_5TT_nocrop.mif and T1_HARDI150_5TT_nocrop_WMvolume_3D_HARDItemplate.nii.gz )
Fig 5.9 ( HARDI150_bvecInverseY_WMvolume_act_0.1M_backtrack.tck )
Fig 5.10 ( HARDI150_bvecInverseY_WMvolume_act_0.1M_backtrack.tck )
[ACT commands]:
dwi2response tournier -fslgrad HARDI150_InverseY.bvecs HARDI150.bvals HARDI150.nii.gz HARDI150_bvecInverseY_response.txt
dwi2fod csd HARDI150.nii.gz HARDI150_bvecInverseY_response.txt HARDI150_bvecInverseY_WMvolume_fod.mif -mask T1_HARDI150_5TT_nocrop_WMvolume_3D_HARDItemplate.nii.gz -fslgrad HARDI150_InverseY.bvecs HARDI150.bvals
tckgen -act T1_HARDI150_5TT_nocrop.mif HARDI150_bvecInverseY_WMvolume_fod.mif HARDI150_bvecInverseY_WMvolume_act_0.1M_backtrack.tck -crop_at_gmwmi -seed_gmwmi T1_HARDI150_5TT_nocrop_gmwmi.mif -backtrack -select 100000
Fig 6.1 ( DTI_2013_fod )
Fig. 6.2 ( mrview : T1, DTI, WM, 5TT image in FreeSurfer space)
Fig 6.3 ( DTI_2013_freesurfer_fod.mif )