Hi, Donald,
Thank you very much for your patient explanation!
In my data2 case, I feel very glad to shall the data, but I can’t find the raw DICOM data in my lab, It seems we have lost the raw DICOM data. Or maybe, we didn’t get the original raw data at the beginning. However, if needed, feel free to get the NIFTI data and corresponding T1 data with the dropbox link below.
In my data1 case, I should not transform DWI data to another space, as you said it will damage the original orientation information. So, I try to warp tck file, the corresponding FA image to MNI152 space.
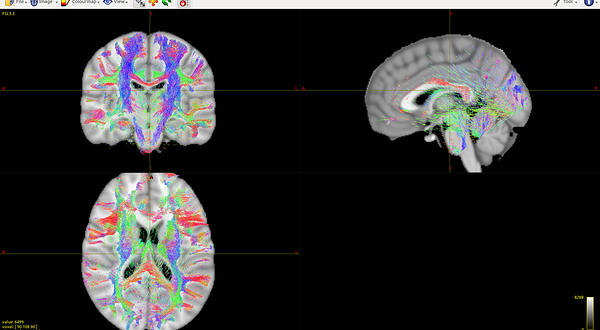
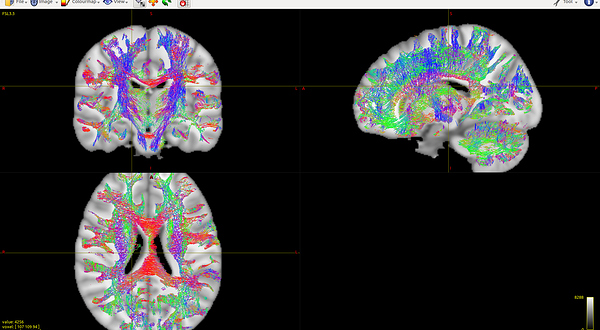
In order to test the warping method, I almost copy Eloy’s steps. All the steps go without any error. However, there are quite a lot straight lines along with the fibers, especially from the sagittal view ( Fig 7.1 , data1), which is quite different from the warped result with the same steps of data3 (Fig 7.2, data3). Are there any restrictive conditions on warping the images? I guess the problem should hide in my data1.
Also, Comparing with the tck file in subject space, the tck file warped to MNI152 space reserved the same streamline numbers and point numbers. However, the length of each streamline changed. Also, the fa value of each point is quite different from the original value in subject space. Is there any method to preserve the original information after warping to MNI152 space?
A lot of Thanks,
Chaoqing
The dropbox link to HARDI150 data:
The warping steps of data 1:
fslchfiletype NIFTI MNI152_T1_1mm.nii.gz
ANTS 3 -m CC[MNI152_T1_1mm.nii, t1_2013.nii.gz,1,5] -t SyN[0.5] -r Gauss[2,0] -o T1_to_MNI_synants.nii -i 30x90x20 --use-Histogram-Matching
warpinit MNI152_T1_1mm.nii flirtMNI-.nii
for i in {0…2}
do
WarpImageMultiTransform 3 flirtMNI-{i}.nii flirtMNI2tck-{i}.nii -R MNI152_T1_1mm.nii -i T1_to_MNI_synantsAffine.txt T1_to_MNI_synantsInverseWarp.nii
donewarpcorrect flirtMNI2tck-.nii flirtMNI2tck_corr.mif
tcknormalise DTI_2013_act_0.5M.tck flirtMNI2tck_corr
.mif DTI_2013_act_0.5M_WarpMNI152.tck
Fig 7.1 ( data1 warped tckfile ):
Fig 7.2 (data3 warped tckfile):
The warping steps of data 3 and FA value sampling.
ANTS 3 -m CC[MNI152_T1_1mm.nii, t1_mpr_ns_sag_iso.nii.gz, 1, 5] -t SyN[0.5] -r Gauss[2,0] -o t1_disease_liuyan_to_MNI_synants.nii -i 30x90x20 --use-Histogram-Matching
warpinit MNI152_T1_1mm.nii flirtMNI-.nii
for i in {0…2}; do WarpImageMultiTransform 3 flirtMNI-{i}.nii flirtMNI2_disease_liuyan_tck-{i}.nii -R MNI152_T1_1mm.nii -i t1_disease_liuyan_to_MNI_synantsAffine.txt t1_disease_liuyan_to_MNI_synantsInverseWarp.nii ; done
warpcorrect flirtMNI2_disease_liuyan_tck-.nii flirtMNI2_disease_liuyan_tck_corr.mif
tcknormalise DTI_30_average-6_act_sift_0.2M.tck flirtMNI2_disease_liuyan_tck_corr.mif DTI_30_average-6_act_sift_0.2M_warpMNI152.tck
dwi2tensor –mask DTI_30_average-6_mask.nii DTI_30_average-6.nii DTI_30_average-6_tensor.nii –fslgrad DTI_30_average-6_bvecs DTI_30_average-6_bvals
tensor2metric –fa DTI_30_average-6_fa.mif DTI_30_average-6_tensor.nii
tcksample -stat_tck mean DTI_30_average-6_act_sift_0.2M_warpMNI152.tck DTI_30_average-6_fa.mif DTI_30_average-6_act_sift_0.2M_warpMNI152_MeanFA.txt