Hi, Donald,
I mainly test ACT method on two data sets, and what I concerned most are the following two points:
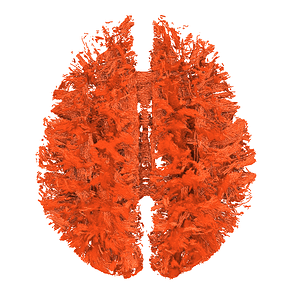
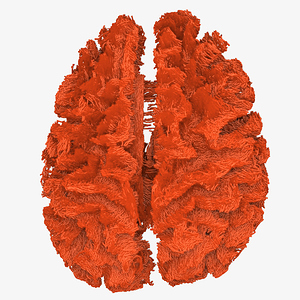
- The corpus callosum fibers are incomplete, as shown in Fig.1-5 and Fig 2-5.
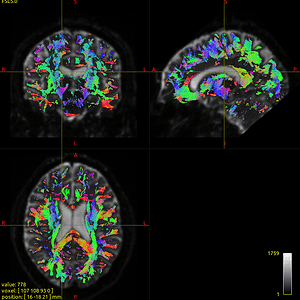
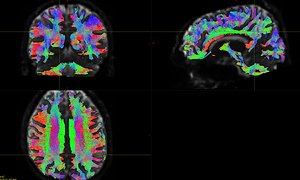
- The tckgen results a lot of short fibers, as shown in Fig 1-4.
I also had tried:
A) increase fiber number: up to 1M;
B) Change mask: white matter mask → whole brain mask.
C) seeding mechanisms: -seed_dynamic and -seed_gmwmi
But the comeout has no any improvement.
Can you provide me some suggestion?
Thanks,
Chaoqing
1. ACT with data1
I register DTI and T1 to MNI152 space as Lucius suggested.
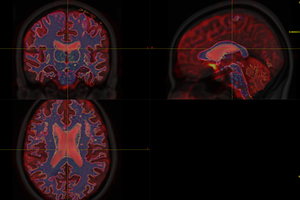
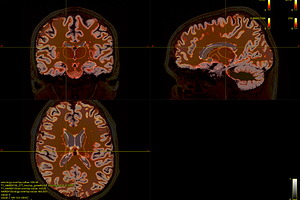
Fig. 1-0. ( mrview: T1, DTI, WM, GMWMI in MNI152 space)
Fig. 1-1. ( response function )
Fig. 1-2. (white matter mask)
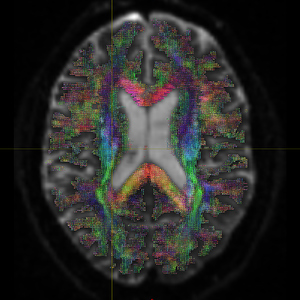
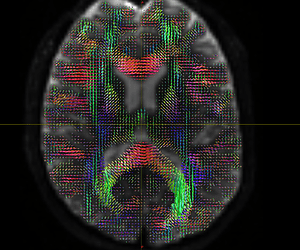
Fig. 1-3. (Fiber orientation distribution)
Fig. 1-4. (tckgen -act result)
Fig. 1-5. (The rendering result)
[ACT commands]:
5ttgen fsl t1_2013_freesufer_in_MNI152.nii.gz t1_2013_freesufer_in_MNI152_nocrop_5TT.nii.gz -nocrop
5tt2gmwmi t1_2013_freesufer_in_MNI152_nocrop_5TT.nii.gz t1_2013_freesufe
r_in_MNI152_nocrop_5TT_gmwmi.mifdwi2response tournier DTI_2013_freesurfer_in_MNI152.nii.gz DTI_2013_free
surfer_in_MNI152_fswm_response.txt -mask t1_2013_freesufer_wm_in_MNI152.nii.gz -fslgrad DTI_2013.bvecs DTI_2013.bvalsdwi2fod csd DTI_2013_freesurfer_in_MNI152.nii.gz DTI_2013_freesurfer_in_
MNI152_fswm_response.txt DTI_2013_freesurfer_in_MNI152_fswm_fod.mif -mask t1_2013_freesufer_wm_in_MNI152.nii.gz -fslgrad DTI_2013.bvecs DTI_2013.bvalstckgen -act t1_2013_freesufer_in_MNI152_nocrop_5TT.nii.gz DTI_2013_free
surfer_in_MNI152_fswm_fod.mif DTI_2013_freesurfer_in_MNI152_fswm_act_gmwmi_backtrack_0.1M.tck -crop_at_gmwmi -seed_gmwmi t1_2013_freesufer_wm_in_MNI152.nii.gz -backtrack -select 100000
2. ACT with data2(An example data)
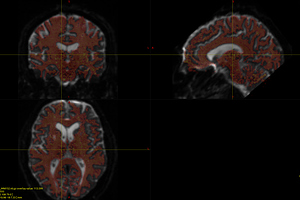
Fig. 2-0 ( mrview: T1, DTI, WM, GMWMI )
Fig. 2-1. ( response function )
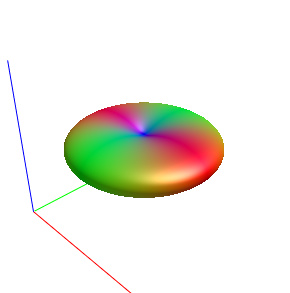
Fig. 2-3. (Fiber orientation distribution)
Fig. 2-4. (tckgen -act result)
Fig. 2-5. (The rendering result)
[ACT commands]:
5ttgen fsl T1_HARDI150.nii.gz T1_HARDI150_5TT_nocrop.mif -nocrop
dwi2response tournier -fslgrad HARDI150.bvec HARDI150.bval HARDI150.nii.gz HARDI150_response.txt
dwi2fod csd HARDI150.nii.gz HARDI150_response.txt HARDI150_WMvolume_fod.mif -mask T1_HARDI150_5TT_nocrop_WMvolume_3D_HARDItemplate.nii.gz -fslgrad HARDI150.bvec HARDI150.bval
5tt2gmwmi T1_HARDI150_5TT_nocrop.mif T1_HARDI150_5TT_nocrop_gmwmi.mif
tckgen -act T1_HARDI150_5TT_nocrop.mif HARDI150_WMvolume_fod.mif HARDI150_Wmvolume_act_0.1M_backtrck.tck -crop_at_gmwmi -seed_gmwmi T1_HARDI150_5TT_nocrop_gmwmi.mif -backtrack -select 100000