Hi all,
I’m working on a dataset which contains two dwis for each subject. Visually, the brain is bigger in one of the acquisitions. I converted them into a single dwi file using mrcat -axis 3 and run dwipreproc after denoising. I expected that all dwi images have been registered to the first b0 after preprocessing and be of the same size but they’re not still the same size after. Interestingly, dwi2mask builds the mask from the larger b0 image. I think this size difference can produce false estimations in further analysis (dwi2tensor, tractography, etc.). FOV for both is 12812864, Slice thickness=2mm. Is there a way that I could be able to correct this issue?
Bests,
Amir
The first thing to figure out is why the brain size differs between the two scans - this simply shouldn’t happen if they were acquired with the same parameters. The simplest explanation is that each was acquired with opposite phase-encode direction for the EPI readout - in which case the apparent difference in size would be due to opposite susceptibility-induced distortions, which you should be able to correct for using the right parameters to dwipreproc. Maybe if you post some screenshots of the problem, we might have a better idea of what the issue is exactly…
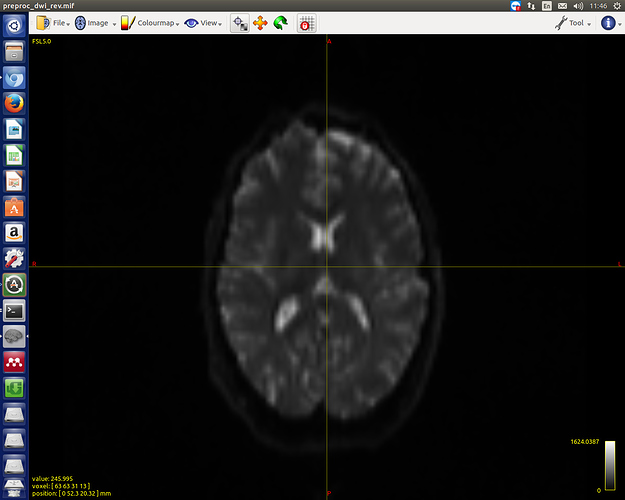
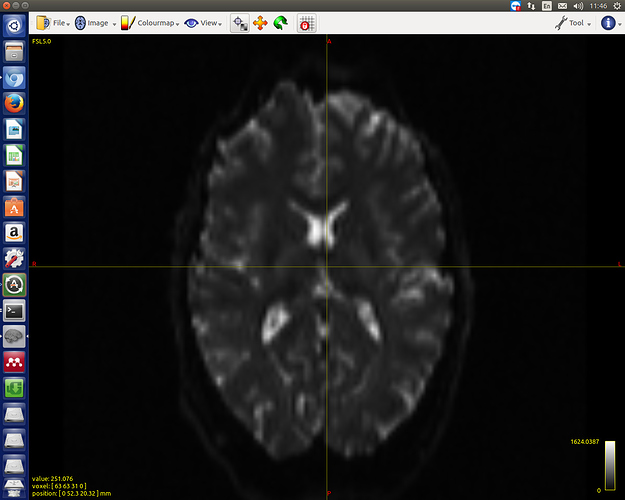
Thanks for the reply. Here are the screenshots of two b0s:
OK, that’s not due to distortions… What does mrinfo report for the two separate images? It looks like they have the same dimensions (otherwise mrcat wouldn’t have worked), so they must have different voxel sizes (and hence different physical FoVs)… If you have access to the original DICOM, you could drill down into the exact acquisition details using dcminfo. But as it stands, there’s no reasonable way to combine both datasets, in my opinion…
Thanks. I checked using mrinfo, one of the images have dimensions of 2.52.52 (the other one is 222). I think it would be rational to choose the 222 one for further analysis.