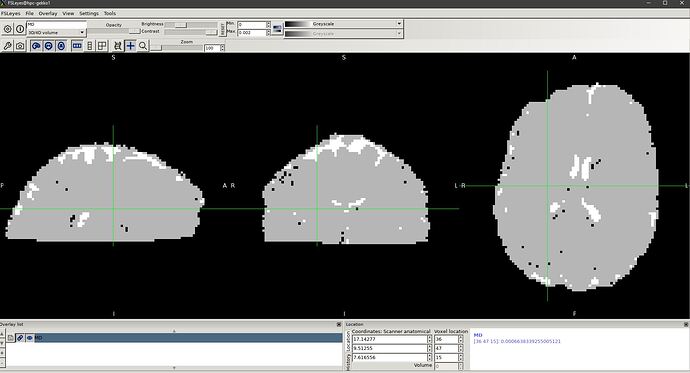
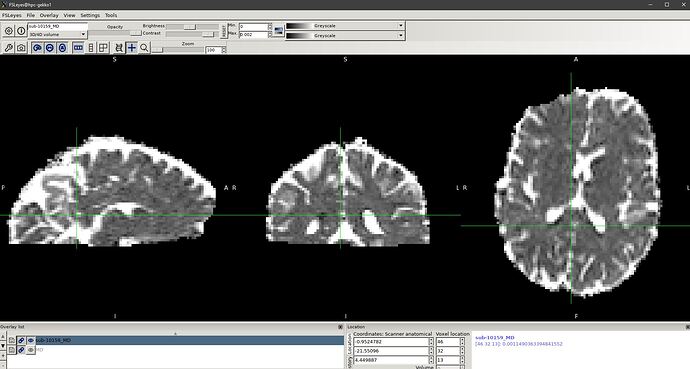
I’ve gotten a pretty normal looking FA image from tensor2metric, however the MD/ADC image looks a little weird (see the screenshot below). If i use FSL dtifit on the preprocessed DTI image, the MD image turns out fine
Yes, I agree that doesn’t look right… Can you post the output of:
mrinfo MD.nii
mrstats MD.nii
assuming your MD image is called MD.nii?
(mrtrix3env) [junhong.yu@hpc-gekko1 sub-10159]$ fsleyes MD.nii.gz
(mrtrix3env) [junhong.yu@hpc-gekko1 sub-10159]$ mrstats MD.nii.gz
mrstats: [100%] uncompressing image "MD.nii.gz"
volume mean median std min max count
[ 0 ] -0.000769833 0 0.299673 -128.054 1.42593 452624
(mrtrix3env) [junhong.yu@hpc-gekko1 sub-10159]$ mrinfo MD.nii.gz
************************************************
Image name: "MD.nii.gz"
************************************************
Dimensions: 96 x 96 x 50
Voxel size: 1.97917 x 1.97917 x 2
Data strides: [ -1 2 3 ]
Format: NIfTI-1.1 (GZip compressed)
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 1 0 -0 -98.88
0 1 -0 -84.5
-0 0 1 -22.38
comments: TE=93;Time=104347
mrtrix_version: 3.0.3
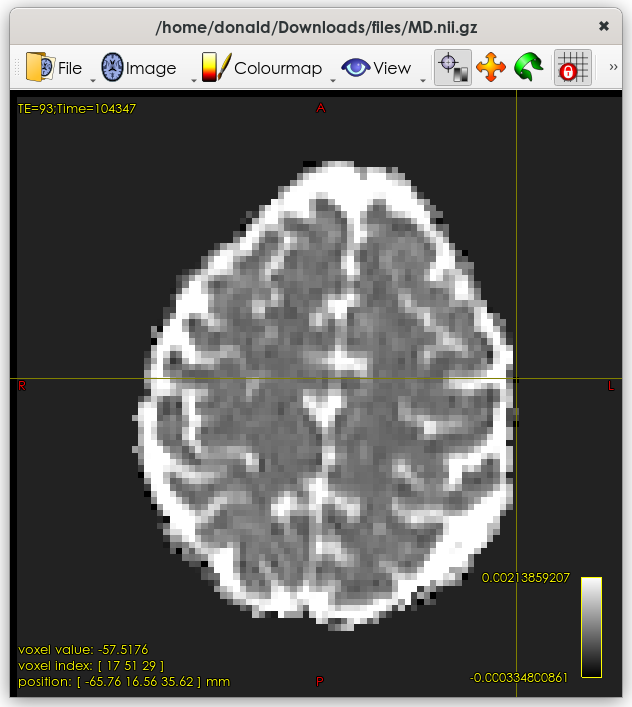
OK, I can see some values that are clearly wrong (MD = -128 is clearly non-physical, and MD = 1.4 is also way too high for the brain). But these values might fall in non-parenchyma regions and might not be relevant anyway. They may however make it difficult to set the right intensity range for the display. I’m not familiar enough with FSLeyes to know how that might affect its display, but setting the range explicitly to [ 0 0.002 ] or so may help. I would also like to know whether you see the same issue when using MRView to display the same data – again setting the range manually if needed (you can use the View tool for that). Any chance you could post a screenshot from MRView as well?
thanks for the quick response. Setting the range to [0 0.002] does not help. I’ve attached 2 screenshots of fsleyes showing the MD image generated from tensor2metric and dtifit
unfortunately, i’m not able to use MRview due to some opengl related issues— i’m waiting for my server admin to update the opengl libraries
OK, rather than waiting for your sysadmin to get OpenGL working (which is no trivial undertaking…), you could alternatively send me the problematic file so I can double-check? You can use any file-sharing service you’re comfortable with, and send to jdtournier@gmail.com.
you can download the file from https://drive.google.com/file/d/19edruKDbFxtrfnYeqLVkULjSPFFYMIwN/view?usp=sharing
i’ve included the tensor file and MD image
thanks
OK, the image is basically fine:
But it does contain some non-sensible values in the odd voxel at the edge of the brain – for example, the value in the voxel under the crosshairs is -57, which is non-physical.
My guess for why fsleyes doesn’t handle these images well is probably because it’s converting these floating-point values to integer representation to then load onto the GPU, which then has trouble handling the range of values present in the image, leading to discretisation errors.
One way to deal with this might be simply to clamp the values in the plausible range, see if fsleyes handles that better. You can do that with a command like this:
mrcalc MD.nii.gz 0 -max 0.01 -min MD_clamped.nii.gz
Hello @jdtournier ,
Thanks for the info.  I would like to know what clamping here means. Does it zero the values greater than the max value or does it keep max value to all the voxels greater than max value?
I would like to know what clamping here means. Does it zero the values greater than the max value or does it keep max value to all the voxels greater than max value?
I guess max is 0.01 and min is 0 in the above command.
Many Thanks,
Suren
What I mean here is that any value lower than 0 gets assigned the value 0, and any value higher than 0.01 gets assigned the value 0.01.
The operation that’s being performed is:
min (max (input_value, 0), 0.01)
And yes, the clamping in the example above was to the range [ 0 0.01 ].
yes i can confirm the clamped MD image looks fine in fsleyes. thanks!