Hi @grahamwjohnson,
For reference, some relevant sources of cautionary information:
- https://community.mrtrix.org/t/mrtrix-tutorial-available-on-osf/1942/32
- https://community.mrtrix.org/t/fiber-column-tracking-in-the-gray-matter/2368/4
- https://community.mrtrix.org/t/fiber-column-tracking-in-the-gray-matter/2368/6
I’ve recently asked someone’s experience with allowing to track into cortical GM by using the current ACT rules / segmentation type for “sub-cortical grey matter”, and they reported what we suspected all along: the precision of a voxel-wise (even with partial voluming / non-binary) segmentation of the cortex isn’t sufficient to represent the correct continuity and/or topology of the outer cortical surface, due to how narrow many sulci are in healthy human subjects. In practice, this means many parts of the outer cortical surface touch / “connect” to those of topologically distinct parts of the cortex. Such anatomical constraints then very easily allow for many false positive assignments (or in general, end points) to entirely wrong parts of the cortex; i.e. they don’t sufficiently constrain streamlines to the correct part of the cortex.
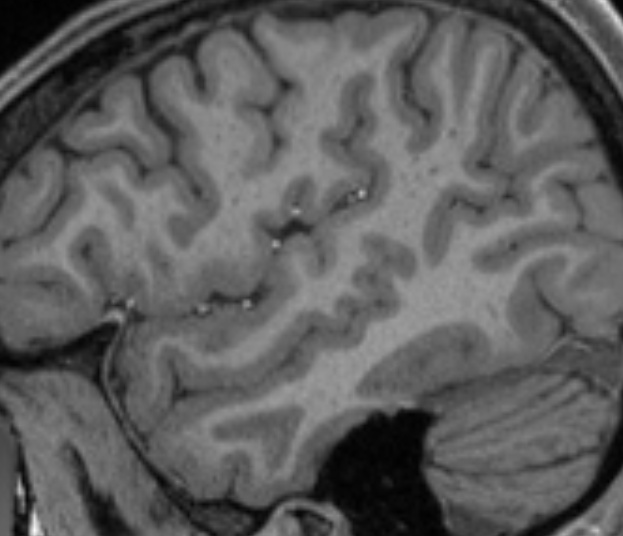
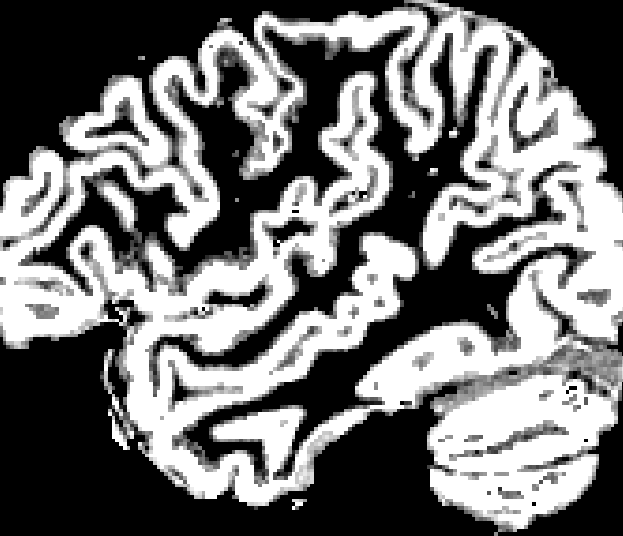
Note this even still holds extensively for e.g the HCP data, which has an outstanding anatomical voxel size of 0.7 x 0.7 x 0.7 mm^3. A quick qualitative inspection of the 5TT GM segmentation reveals this:
T1w image:
5TT GM segmentation, with partial volume:
Good examples of the problem can be seen along the entire length of e.g. the superior temporal sulcus (…at least I think that’s what it’s called), but you’ll easily spot many other locations as well, even only in this screenshot.
Don’t get me wrong; there’s nothing wrong with the segmentation: it’s done a very reasonable job on the anatomical image above for sure. But the precision due to partial voluming simply falls short to represent the topology of the outer cortical surface in a reliable way. Consequentially, you can’t rely on such a segmentation combined with a rule set that relies on a robust outer cortical surface identification. I’ve been providing some people with a few tips and tricks to pragmatically deal with some of this, but the fundamental limitation itself is already introduced at this stage: some of the lost precision can simply not be recovered once lost. A mesh segmentation of both inner and outer surfaces of the cortex allows to retain the required precision, while imposing solid topological assumptions on the segmentation process itself.
So in any case, take that into account in terms of what precision you can expect from such a process. It might not help much currently, but it’s an relevant reality check.
Cheers,
Thijs