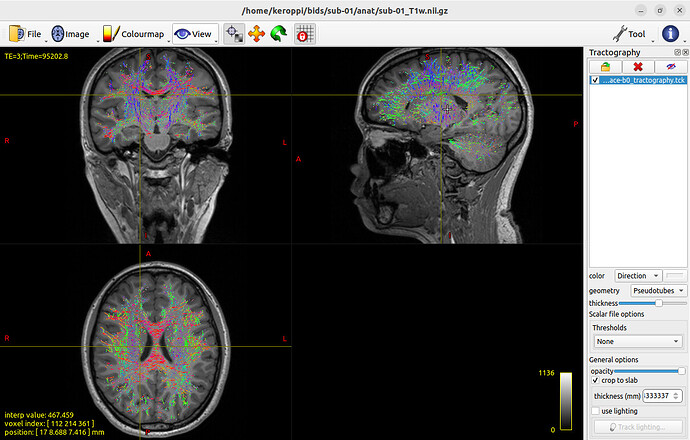
Hi to mrtrixers!
I am working on 2 human groups’ brain tractography. But the .tck files seem weird because the fiber lengths are much shorter than what I expected.
My DWI data for both subject groups underwent eddy only. As single-shell data, I ran:
-
dwi2response tournier -shells 1000,0 -nthreads 8
and averaged responses within groups (responsemean command). -
FODs were calculated via:
dwi2fod -shells 1000 msmt_csd dwi_eddy_prepro.nii common_mean_res.txt “$fod_file” -mask dwi_eddy_prepro_mask.nii. -
5ttgen T1_raw.mif 5tt_nocoreg.mif
-
flirt T1_raw.nii mean_b0_dwi_eddy_prepro.mif -dof 6 -omat diff2struct_fsl.mat
-
transformconvert diff2struct_fsl.mat mean_b0_dwi_eddy_prepro.mif T1_raw.mif flirt_import diff2struct_mrtrix.txt
-
Coregistered T1_coreg.mif and 5tt_coreg.mif via:
mrtransform -linear diff2struct_mrtrix.txt -inverse. -
Generated GM-WM interface mask:
5tt2gmwmi 5tt_coreg.mif gmwmSeed_coreg.mif. -
Executed tractography:
tckgen -act 5tt_coreg.mif -backtrack -seed_gmwmi gmwmSeed_coreg.mif -select 10000 wmfod_norm.mif tracks_10k.tck.
Questions:
- Are my parameters and steps reasonable?
- Are abnormally short whole-brain tracts (see fig.1) caused by inadequate preprocessing (e.g., missing bias field correction)?
Many thanks
Yaqin