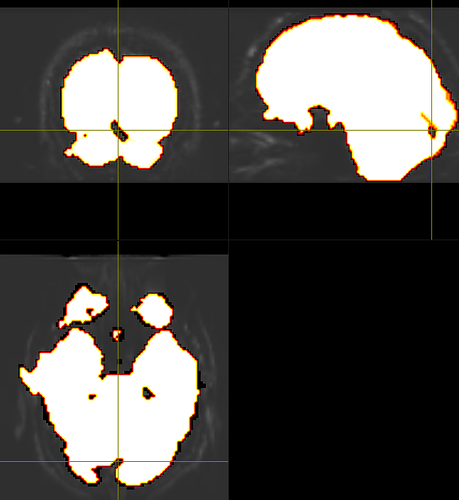
And my output, upon running dwibiascorrect command, I get this:
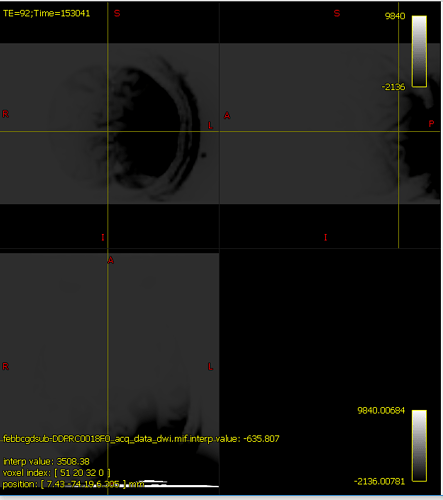
It’s hard to know exactly what’s going on here because mrview is not just displaying one image, there’s an overlay image being placed on top; and if the overlay opacity is 1.0, that can result in the overlayed image hiding the main image, even if it’s filled with zeroes. In your case, the intensity windowing on the overlaid image means that pretty much any reasonable image would come out as grey.
Better would be to show the input and output images with the same intensity windowing, without ever touching the overlay tool, as well as the output of mrstats on both.
Actually on second thought, I think maybe I know what’s going on here:
-
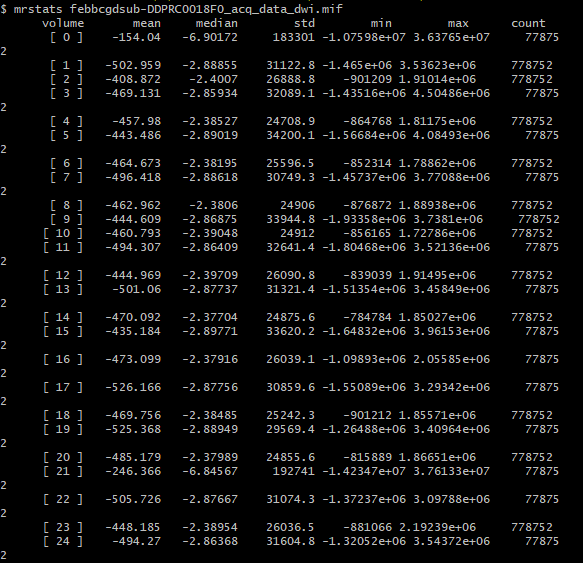
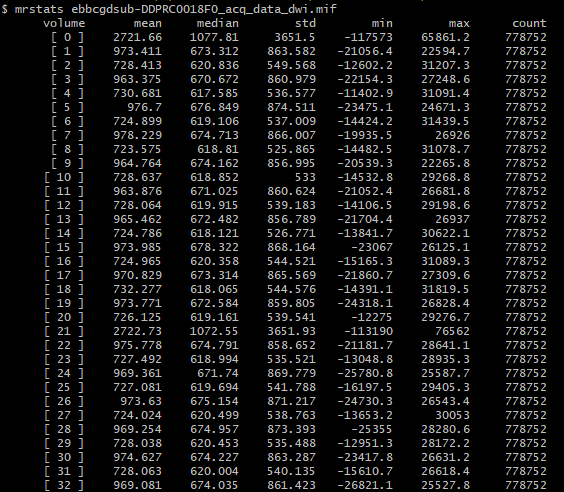
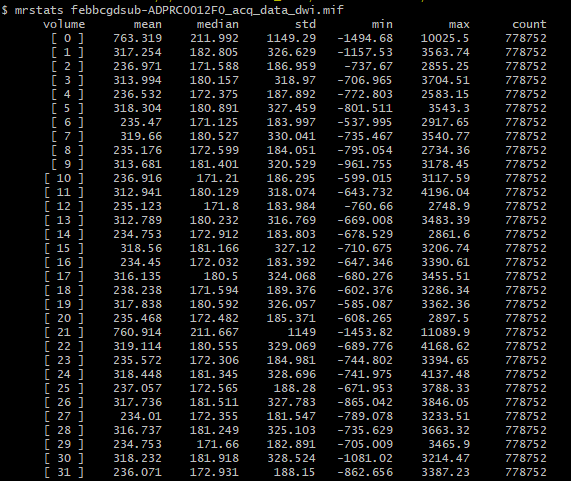
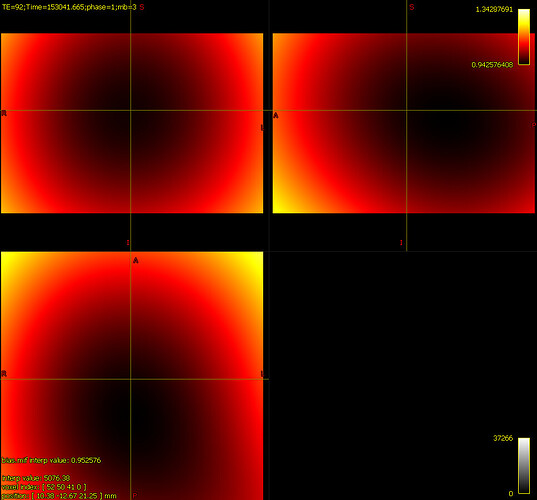
There’s extreme values hiding somewhere in your DWI volume.
-
When you load the original DWI volume as the main image in mrview, the intensity windowing is determined based on the maximum and minimum intensities in the first slice you view.
-
When you load the bias-field-corrected image in the overlay tool in mrview, the intensity windowing is determined based on the maximum and minimum intensities of the whole volume (as there’s no guarantee that an overlay image resides on the same voxel grid as the main image, it could be in any arbitrary orientation, and therefore the whole volume needs to be checked in order to determine appropriate windowing).
The colour bar at the top right of the window is the intensity scaling of the overlay image. With a range of -10,000,000 to 36,000,000, almost any reasonable neuroimaging data is going to appear as a flat dark grey. Even the whitest areas of whatever image is loaded as the main image, with intensity 37,000, are sufficiently close to 0.0 with your current windowing to be indistinguishable.
There are two possibilities. Either these extreme values are actually present in your input DWI data and you just didn’t notice because of the way mrview determines its intensity windowing, or dwibiascorrect has somehow introduced those extreme values in between producing “result.mif” and mrconverting that image to your requested output image location. Or maybe it’s there in “result.mif” too, but you manually altered the intensity windowing in that case?
In general, robust masking is proving remarkably difficult to achieve consistently, as @rsmith will no doubt confirm…
If there were an appropriate emoji for PTSD I would use it ad nauseum.
Nevertheless, if you’re either interested in or frustrated with DWI brain masking, you may be interested in these changes coming for 3.1.0, which provides many algorithms to choose from.
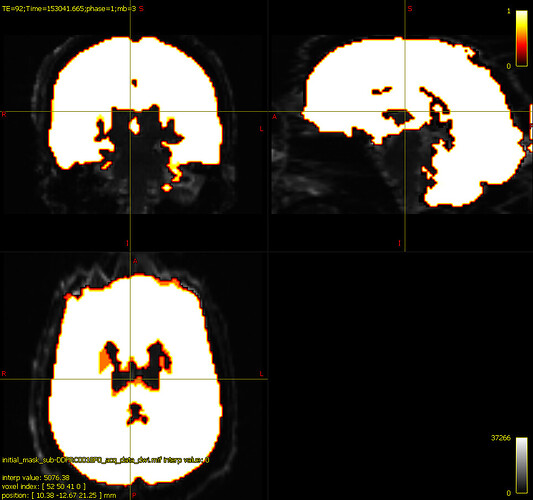
one of my colleagues is using the ‘erode’ option from mrcalc to constrain the BET mask more
maskfilter* 
would it be better to use a second bias corrected image from the generation from the second mask? The second mask tends to look more ‘wholesome’ like the one above, but when comparing the bias corrected images, they don’t seem to reveal too many differences
Theoretically, if you were to run some large number of iterations where you bounce between mask determination and bias field estimation, after some number of iterations there should no longer be any difference between subsequent iterations. Whether or not this is the case however depends on the precision of whatever is providing the bias field estimates, and the robustness of the brain masking algorithm. And how many iterations would be required depends on the nuances of those algorithms also.
This is an idea that I’ve actually been tinkering with myself for quite some time: iterating between these and measuring differences in binary masks between iterations to detect convergence. It can work, but it’s still constrained by the limitations of whatever brain masking algorithm is being utilised.
What I’ve settled on myself for now for my own DWI pre-processing tasks (adjusts tie ahead of shameless self-plug) is in the 0.5.0 update of my connectome BIDS App. My approach is further different (code for anyone so inclined) in that I’m utilising mtnormalise to both bias field correct the DWIs and provide a total tissue density image that I then threshold to produce a binary brain mask (on the assumption that a brain mask should include voxels where there are brain tissues or fluid and not those where there is not), which is then fed back to the start of the iterative loop. In this specific instance iterating repeatedly can be detrimental in some data because of mtnormalise’s sensitivity to the inclusion of non-brain voxels in the mask, which self-perpetuates if performing iteratively, hence why I only iterate twice for this particular approach; but when I was using just dwi2mask in the same loop I could get something approaching convergence after 5-10 iterations.
Maybe I should convert that approach into a dwi2mask algorithm…
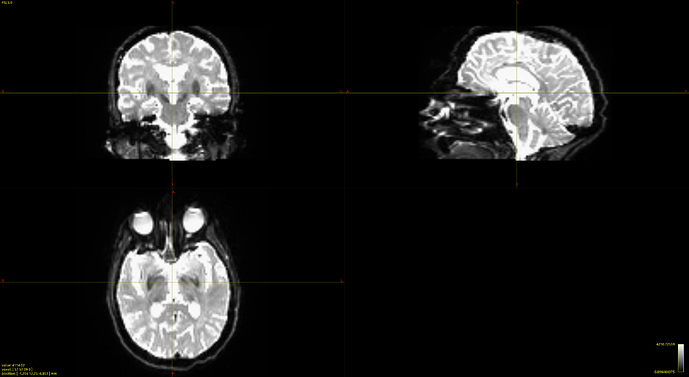
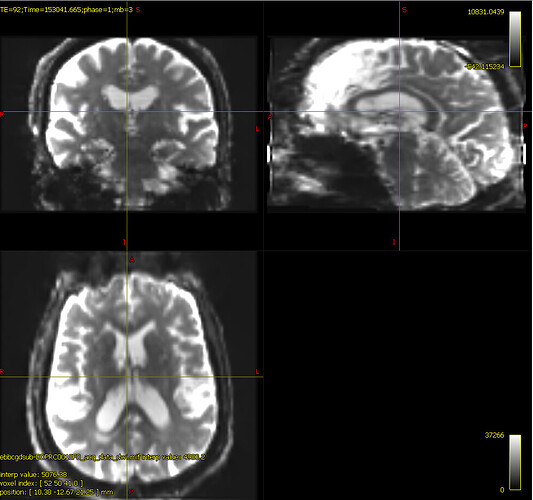

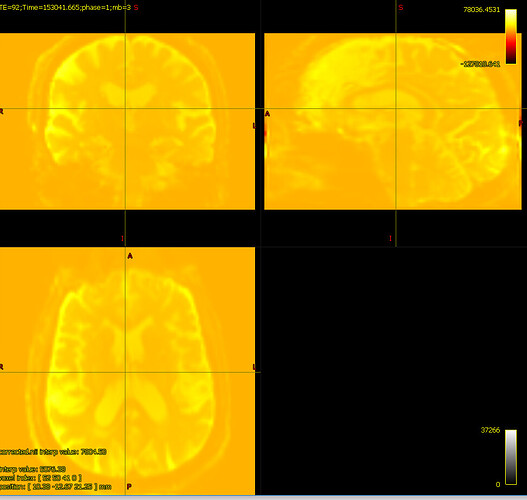
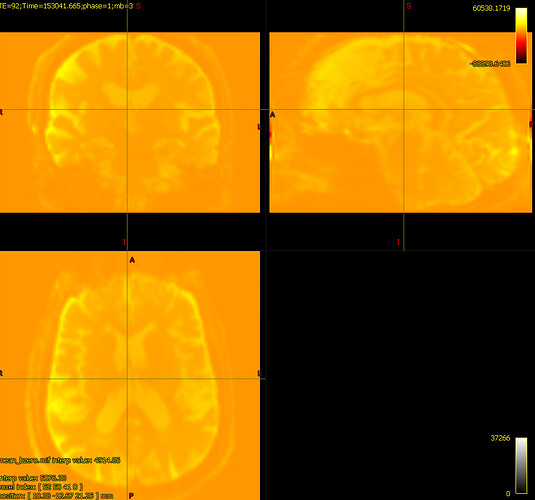


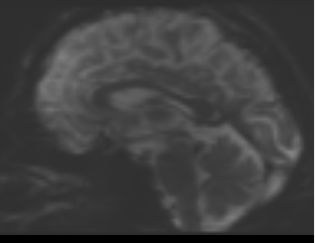 bias corrected image with first pass mask
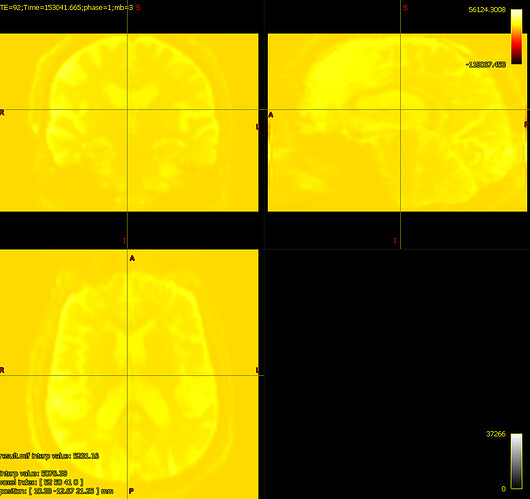
bias corrected image with first pass mask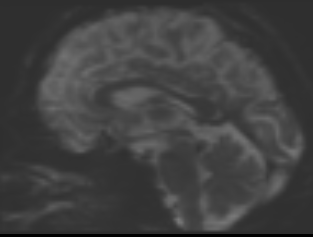 bias corrected image with second pass mask
bias corrected image with second pass mask