Dear Mrtrix Team,
When I run the pipeline https://mrtrix.readthedocs.io/en/latest/fixel_based_analysis/mt_fibre_density_cross-section.html I got an error message at step 8/9 before/when the population template supposed to be calculated:
ln: illegal option – r
usage: ln [-Ffhinsv] source_file [target_file]
ln [-Ffhinsv] source_file … target_dir
link source_file target_file
ln: illegal option – r
usage: ln [-Ffhinsv] source_file [target_file]
ln [-Ffhinsv] source_file … target_dir
link source_file target_file
Error: unrecognized arguments: -voxel_size 1.25
Usage: population_template input_dir template [ options ]
(Run population_template -help for more information)
this are my commands (of my bash script loop):
mtnormalise ${dir}${tp}/wmfod.mif ${dir}${tp}/wmfod_norm.mif ${dir}${tp}/gm.mif ${dir}${tp}/gm_norm.mif ${dir}${tp}/csf.mif ${dir}${tp}/csf_norm.mif -mask ${dir}${tp}/${dir}${tp}_mask_upsampled.nii -debug -force
ln -sr ${dir}${tp}/wmfod_norm.mif $dir/fod_input/${dir}${tp}.mif ; ln -sr ${dir}${tp}/${dir}${tp}_mask_upsampled.nii $dir/mask_input/${dir}${tp}.nii
population_template $dir/fod_input -mask_dir $dir/mask_input $dir/${dir}_wmfod_template.mif -voxel_size 1.25 -type rigid -debug -force
Thank you for your support!
Best
max
Hi @Max_Wichmann,
That’s probably due to your MRtrix3 installation not being quite up to date. @maxpietsch added the -voxel_size option–I think–sometime during the development of either RC2 or RC3 (the most recent versions of MRtrix3). I’d strongly urge you to update to the latest RC3 version in any case if you’re going through the steps for a fixel based analysis: an important bug in tckgen was fixed in RC3, which directly (and significantly) affects the advice in the whole brain tractography step in the fixel-based analysis pipeline (the documentation you’re referring to).
Cheers,
Thijs
Alright ! Thank you for this advice! It works now!
However, while it calculated the population_template I got this new Error message:
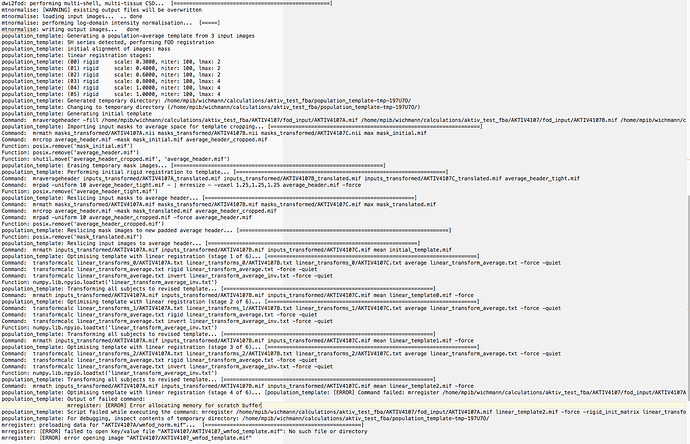
mrregister ./fod_input/AKTIV2240A.mif linear_template2.mif -force -rigid_init_matrix linear_transforms_2/AKTIV2240A.txt -mask1 ./AKTIV2240/mask_input/AKTIV2240A.nii -rigid_scale 0.8 -rigid_niter 100 -rigid_lmax 4 -type rigid -datatype float32 -rigid linear_transforms_3/AKTIV2240A.txt
mrregister: [ERROR] Error allocating memory for scratch buffer
Here is an image of the debugging:
I do the processing on a Grid-Server (cluster).
I already checked this post, but I do not really understand the solution: http://community.mrtrix.org/t/mrregister-error-error-allocating-memory-for-scratch-buffer-while-executing-population-template-command/403
Thank you for any kind of help!
Best
Max
The post you link to is from quite a while ago. I’m not 100% on top of all our changes related to the various temporary directories we use since… so not sure whether that post still applies exactly.
@jdtournier @maxpietsch , maybe you guys can help out with this one…?
Thank you for your help, I guess I solve the problem. I had to change the RAM options inside the job file which I have to send to the cluster.
Thank you
Max