Dear MRtrix3 experts,
I’m running the dwidenoise and dwipreproc commands on a data set and appear to be running into problems.
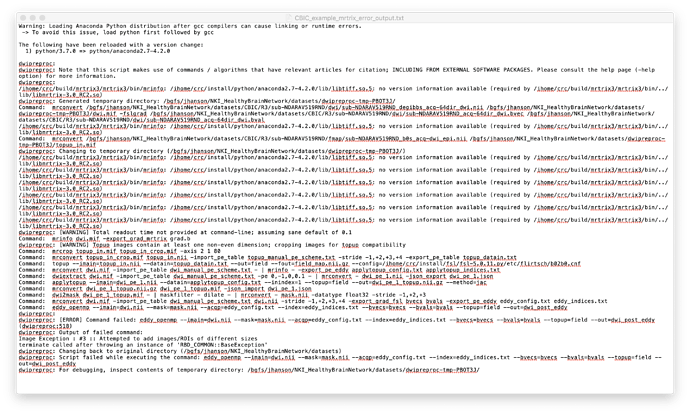
The first is with dwipreproc:
$ dwipreproc -nthreads 20 dwi_denoised.mif dwi_preprocessed.mif -pe_dir AP -rpe_pair -se_epi b0s.mif
dwipreproc:
dwipreproc: Note that this script makes use of commands / algorithms that have relevant articles for citation; INCLUDING FROM EXTERNAL SOFTWARE PACKAGES. Please consult the help page (-help option) for more information.
dwipreproc:
dwipreproc: Generated temporary directory: /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwipreproc-tmp-TNDEA2/
Command: mrconvert /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwi_denoised.mif /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwipreproc-tmp-TNDEA2/dwi.mif
Command: mrconvert /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/b0s.mif /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwipreproc-tmp-TNDEA2/topup_in.mif
dwipreproc: Changing to temporary directory (/ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwipreproc-tmp-TNDEA2/)
Command: mrinfo dwi.mif -export_grad_mrtrix grad.b
dwipreproc: [WARNING] Topup images contain at least one non-even dimension; cropping images for topup compatibility
Command: mrcrop topup_in.mif topup_in_crop.mif -axis 2 1 54
Command: mrconvert topup_in_crop.mif topup_in.nii -import_pe_table topup_manual_pe_scheme.txt -stride -1,+2,+3,+4 -export_pe_table topup_datain.txt
Command: topup --imain=topup_in.nii --datain=topup_datain.txt --out=field --fout=field_map.nii.gz --config=/clusterdata/apps/fsl/etc/flirtsch/b02b0.cnf
Command: mrinfo dwi.mif -export_pe_eddy applytopup_config.txt applytopup_indices.txt
Command: dwiextract dwi.mif -pe 0,-1,0,0.0464 dwi_pe_1.nii
Command: applytopup --imain=dwi_pe_1.nii --datain=applytopup_config.txt --inindex=1 --topup=field --out=dwi_post_topup.nii.gz --method=jac
Command: mrconvert dwi_post_topup.nii.gz -grad grad.b - | dwi2mask - - | maskfilter - dilate - | mrconvert - mask.nii -datatype float32 -stride -1,+2,+3
Command: mrconvert dwi.mif dwi.nii -stride -1,+2,+3,+4 -export_grad_fsl bvecs bvals -export_pe_eddy eddy_config.txt eddy_indices.txt
Command: eddy_openmp --imain=dwi.nii --mask=mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --out=dwi_post_eddy
dwipreproc:
> dwipreproc: [ERROR] Command failed: eddy_openmp --imain=dwi.nii --mask=mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --out=dwi_post_eddy (dwipreproc:470)
> dwipreproc: Output of failed command:
> Image Exception : #3 :: Attempted to add images/ROIs of different sizes
> terminate called after throwing an instance of ‘RBD_COMMON::BaseException’
dwipreproc: Changing back to original directory (/ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI)
dwipreproc: Script failed while executing the command: eddy_openmp --imain=dwi.nii --mask=mask.nii --acqp=eddy_config.txt --index=eddy_indices.txt --bvecs=bvecs --bvals=bvals --topup=field --out=dwi_post_eddy
dwipreproc: For debugging, inspect contents of temporary directory: /ibscratch/users/uqisaham/CAI_imaging/files_relevant_to_analysis/mrtrix3_Preprocessed_Data/S06_KC-3T/HARDI/dwipreproc-tmp-TNDEA2/
The second is with dwidenoise:
dwidenoise ../rawdata/DTI_66/ dwi_denoised.mif dwidenoise: [done] scanning DICOM folder "../rawdata/DTI_66/" dwidenoise: [100%] reading DICOM series "NODDI_OUTER_SHELL_66" dwidenoise: [100%] preloading data for "C_S01_PZ_3T (C_S01_PZ_3T) [MR] NODDI_OUTER_SHELL_66" dwidenoise: [100%] running MP-PCA denoising [uqisaham@inode HARDI] ls
dwi_denoised.mif
[uqisaham@inode HARDI]$ mrview dwi_denoised.mif
mrview: WARNING: invalid key "EchoTime in generic image header “./dwi_denoised.mif” - ignored
mrview: WARNING: invalid key "PhaseEncodingDirection in generic image header “./dwi_denoised.mif” - ignored
mrview: WARNING: invalid key "TotalReadoutTime in generic image header “./dwi_denoised.mif” - ignored
mrview: WARNING: invalid key "command_history in generic image header “./dwi_denoised.mif” - ignored
mrview: WARNING: invalid key "mrtrix_version in generic image header “./dwi_denoised.mif” - ignored
I understand that this issue is currently being resolved and I’m not sure if it relates to the error I receive via dwipreproc.
I’m also aware of this issue with mrview however I’ve been using the same version of MRtrix3 throughout and NIfTI files open without such errors, so not sure how this file corruption was introduced.
To my knowledge, our computers have had a system update recently, however this was a few weeks ago where our IT team confirmed that MRtrix3 was up to date. I will check this with them again tomorrow.
For reference, please find the content check of the mif image:
$ head dwi_denoised.mif
mrtrix image
dim: 100,100,55,67
vox: 2.3,2.3,2.3,nan
layout: -1,-2,+3,+0
datatype: Float32LE
transform: 0.9997041757060217, -0.01797351600319981, -0.01638639064671639, -107.1678526805917
transform: 0.01282302615547002, 0.9619745064493196, -0.272838081912706, -88.60715532724285
transform: 0.02066714968640003, 0.2725472466638971, 0.9619204058859386, -80.40929179312866
EchoTime: 0.078
PhaseEncodingDirection: j-
Your advice on these matters is appreciated!
Ishani