Dear MRtrix developers
I have been trying to run fixel based analysis on mouse DWI sample.
I followed the following site
https://mrtrix.readthedocs.io/en/0.3.16/workflows/fixel_based_analysis.html
I have encountered two issues. First .msf file extension does not support. Therefore, fod2fixel produces fixel file along with index and directions files in a directory. Is there any plan to update the instructions given in the aforementioned website?
I also confused how to organise my files while running Fixelcfestats. Is is a folder containing all the individual samples folders/individual file?
I totally lost?
I will appreciate if you please help me in this instance.
Kind regards
Mamun
Queensland Brain Institute
University of Queensland
Brisbane
It looks like you’re looking at the version of the documentation for 0.3.16. Try the most recent one: click on the bottom bar on the left, you’ll see all available versions of the documentation (they should match the different tags that we’ve released).
In your case, it sounds like you’re after this one. There’s instructions for single-tissue or multi-tissue analysis (though they are mostly identical).
Hi Donald
Thank you very much. Would you please guide me visualising the result?
To visualise the results in step 17 at here
Instructions:
To view the results load the population FOD template image in mrview , and overlay the fixel images using the vector plot tool. Note that p-value images are saved as 1-p-value. Therefore to visualise all p-values < 0.05, threshold the fixels using the vector plot tool at 0.95.
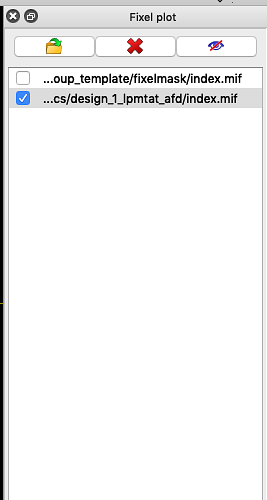
Should I overlay the fixel image (fwe_pvalue.mif) using the vector plot tool (Fixel plot tool)?
How do I select threshold? This option (threshold) is not available in fixel plot tool.
Thanks once again
Kind regards
Mamun
Hi Mamum,
Indeed you have to use the fixelplot.
When you chose which image to color by (eg. t-value) you can then chose which image to threshold with (eg. fwe_pvalue.mif at .95) as illustrated in the image below.
Hi Jeroen
Thank you so much.
I appreciate your help.
Kind regards
Mamun
Hi Donald,
Could you please advise whether I need to change anything (except for the upsampling step) if I wanted to apply this pipeline to the mouse brain?
Thanks
Chiara
Dear Mamun,
I am currently trying to run a fixel based analysis on mice. Could you advice on whether you made any adjustments when running your analysis, or you just followed the standard pipeline?
Thanks
Chiara
Yes, I followed standard pipeline. However, some commands are unavailable in the current Matrix version. For example; you will not find fixelcalc. You may use mrcalc instead. Good luck.
Kind reards
Mamun