Hi Stella,
I also try to use AAL atlas and I want to build the connectivity matrix based on AAL atlas.
To register AAL atlas into subject’s T1 image, I used FSL flirt command.
Here, I described what I tried.
Step 1:
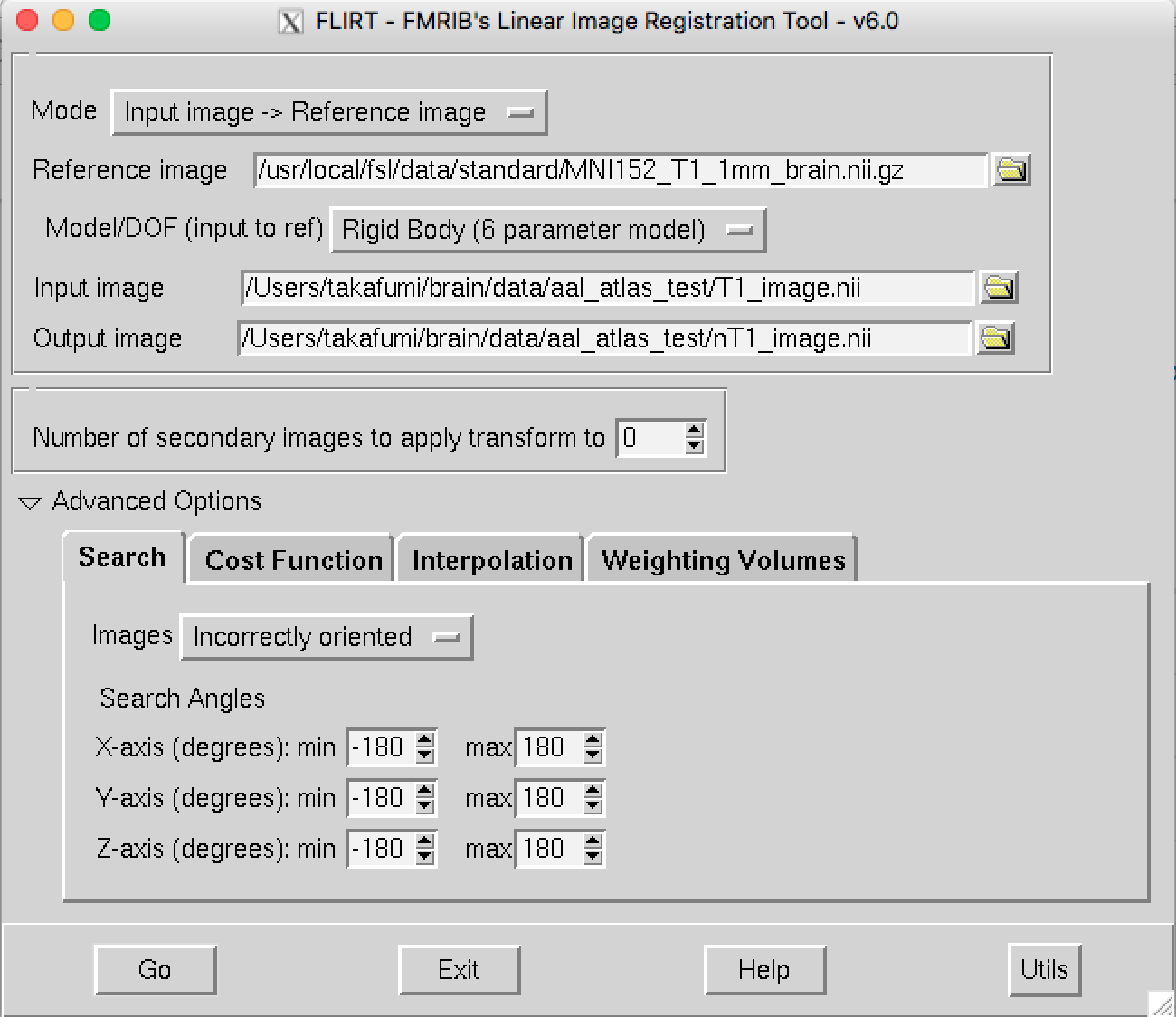
Regiser the subject’s T1 image into MNI template.
By using the FSL FLIRT GUI program, the confgiuration is such that.
Step 2:
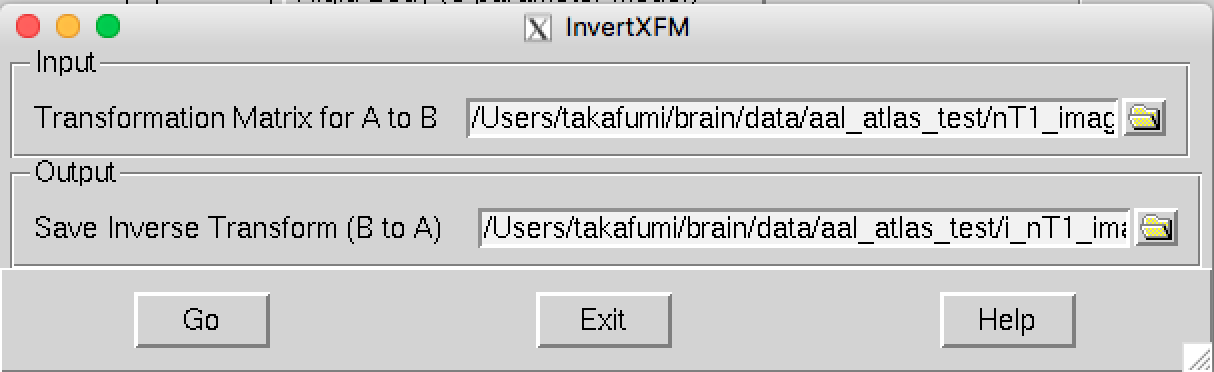
Making the inverse transformation matrix
Step 3:
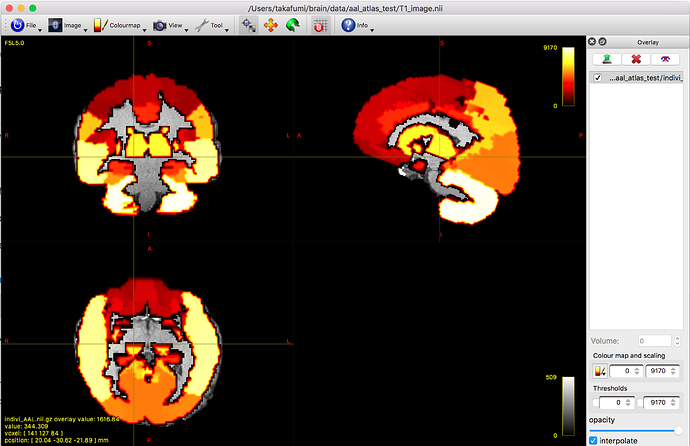
Applying the inverse transformation matrix for AAL atlas and I obtained the AAL atlas on subject space.
Step 4:
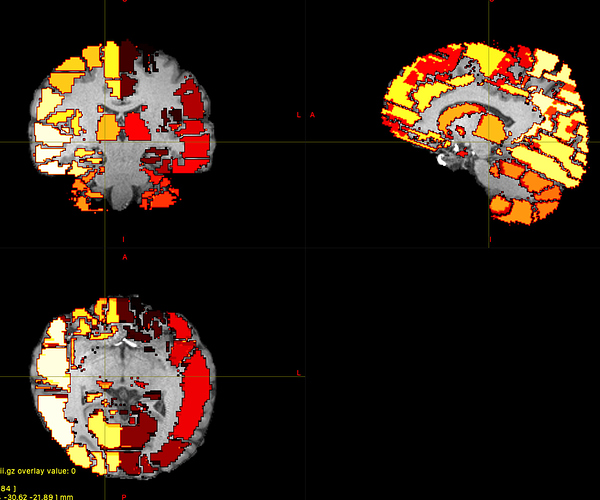
Next, I created the LUT file. and I executted the labelconvert command.
But unfortunately, the obtained AAL atlas was discontinuous.
The original value of AAL file was continuous, then I think this error was caused.
If anyone know the solution to overcome this error, please explain it.
labelconvert indivi_AAL.nii.gz AAL_mapping.txt aal.txt indivi_AAL_fix.nii.gz
Dropbox - File Deleted - Simplify your life
Dropbox - File Deleted - Simplify your life
Thank you!
Cheers.
Takafumi.