Hi all
I am attempting to use tckresample and tcksample to resample FA values along my anterior forceps tract perpendicular to the tangent of an arc. I have followed instructions from a few other posts including this one: Along tract analysis. My commands are:
tckresample genu.tck -arc 20 -18,54,2 0,28,2 18,54,2 genu_samples.tck -nthreads 0
tcksample genu_samples.tck IITmean_FA.nii.gz genu_fa_values.txt
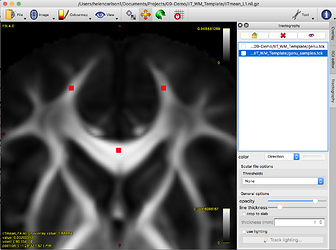
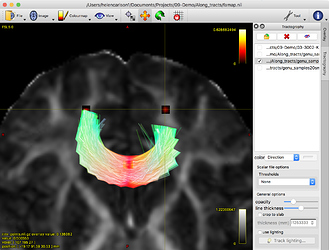
Where the coordinates roughly correspond to the red dots in this image:
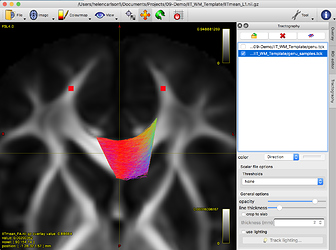
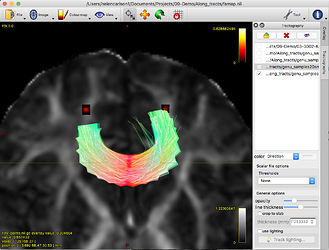
The resulting resampled genu_samples.tck file looks like this:
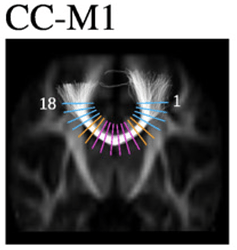
I was expecting something that looks more like this that shows where the samples are located and their angles along the tangent of the tract (an illustration from Groeschel et al, 2014) so that I could verify that I have sufficiently sampled the tract evenly:
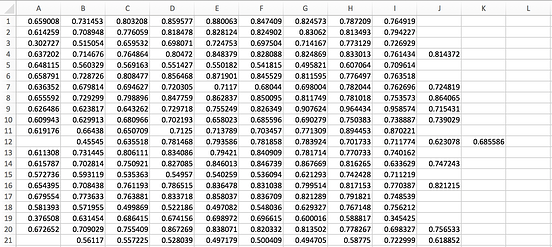
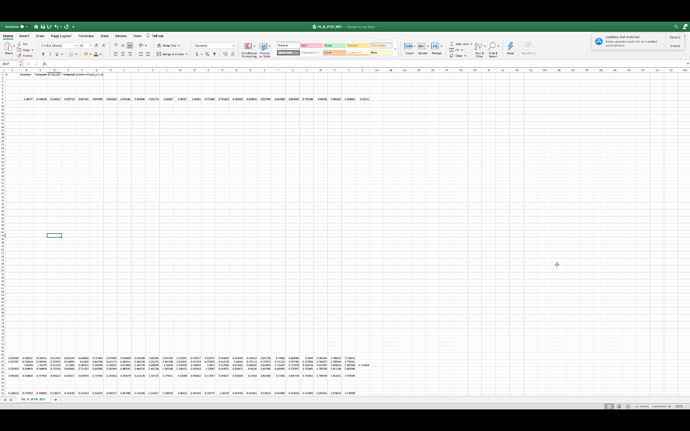
So my question is, how can I be sure that the sampling has come from the entire anterior forceps rather than just the central portion shown in the genu_samples.tck file? The output genu_fa_values.txt file appears to have the correct amount of data in it (20 columns and 4450 rows corresponding to the number of streamlines).
I get a similar result if I move the anterior points closer to the centre point essentially making a smaller arc with shorter streamlines.
Thank-you
Helen