Dear MRtrix-Experts,
I get the 5TT image in T1 space.
Next I transform the 5TT image to DWI space(I have read this post).
flirt -in T1.nii -ref FA.nii -out T12FA.nii -omat T12FA.mat
transformconvert T12FA.mat T1.nii FA.nii flirt_import T12FAmrt.mat
mrtransform 5TT.nii 5TT2FA.nii -linear T12FAmrt.mat
mrtransform: [WARNING] the input linear transform is not orthonormal and therefore applying this without the -template option will mean the output header transform will also be not orthonormal
I find that the qform and sform of the 5TT-coregistered-DWI image are inconsistent
fslhd 5TT2FA.nii
qform_name Scanner Anat
qform_code 1
qto_xyz:1 0.997192 0.005625 0.074613 -104.013123
qto_xyz:2 -0.001768 0.998663 -0.051654 -133.454071
qto_xyz:3 -0.074803 0.051378 0.995874 -67.540840
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 1.030852 0.003590 0.075165 -104.013123
sto_xyz:2 -0.003884 1.021482 -0.058058 -133.454071
sto_xyz:3 -0.075895 0.046108 1.027731 -67.540840
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
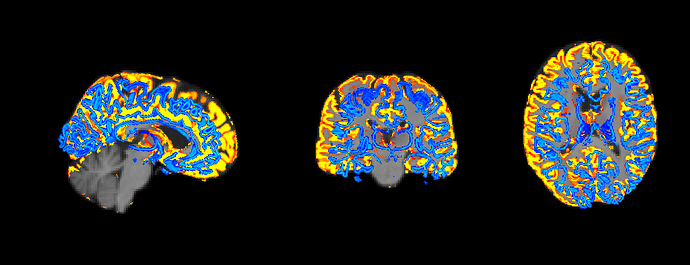
Then using the 5TT-coregistered-DWI image gets gmwmi image
5tt2gmwmi 5TT2FA.nii gmwmi.nii
5tt2gmwmi: [WARNING] voxel spacings inconsistent between NIFTI s-form and header field pixdim
5tt2gmwmi: [WARNING] qform and sform are inconsistent in NIfTI image "5TT2FA.nii" - using qform
5tt2gmwmi: [100%] Generating GMWMI seed mask
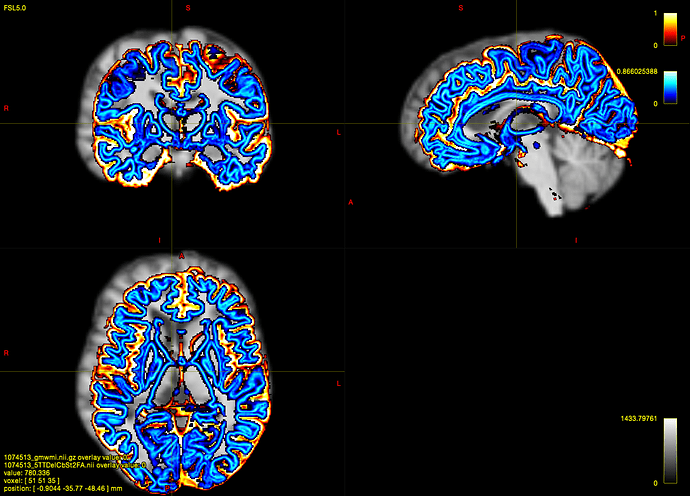
To check the results,I use fsleyes and mrview to test whether they match well.
In fsleyes , the 5TT-coregistered-DWI image and T1-coregistered-DWI image matches well,but the gmwmi image mismatch.
In mrview , the 5TT-coregistered-DWI image and gmwmi image matches well,but the T1-coregistered-DWI image mismatch.!
So My questions are :
1.when transforming 5TT image to DWI space (no regriding ),why sform and qform are inconsistent?
2.The looking like mismatch of the three images in fsleyes or mrview will cause problems about the following analysis such as tractogram or constructing connectome?
Cheers
Liyuan