I wanted to report an issue that I’ve come across while working with HCP data and MRtrix3.
The Human Connectome Project has high quality dMRI data which makes it very appealing to use for tractography studies. Unfortunately, due to the way these data are preprocessed, performing tractography on HCP data with anatomically constrained tractography (ACT) can give some unwanted results.
One such issue that can arise, which has not to my knowledge been reported, is due to the fact that the HCP diffusion data are rigidly transformed to their ACPC aligned space. A by-product of this processing step is that the brainstem may be truncated before reaching the edge of the field of view (FoV). I will demonstrate the problem below.
The data I used was the standard minimally preprocessed HCP data (this example is with participant 100206 ). I will provide the commands used so that anyone else can replicate the error (and potential fixes).
The Problem
As I already mentioned, in the minimally preprocessed HCP data, the DWI volumes are rigidly transformed to the anatomical space, rather than the usual practice of registering the T1w image to the DWI volumes. During this transformation, any voxel in the target image for which there is not a valid location in the input image FoV from which to “pull” image intensities will be filled with zeroes. As a result, the dwi2fod command will populate such voxels with FODs consisting of zeroed coefficients (regardless of whether or not a mask is explicitly provided to dwi2fod ).
This has important ramifications for the inferior part of the brainstem in the context of ACT. While in the acquired DWI data there will be valid image intensities all the way to the inferior-most slice of the image FoV, upon transformation to the ACPC space, this “lower edge” of the brainstem (as defined by the diffusion MRI FoV) will no longer lie at the outer edge of the image FoV. Thus, when a streamline traverses in the inferior direction in this region, it will eventually encounter those voxels where the FODs have zero amplitude, and thus terminate within the brainstem due to the tckgen FOD amplitude threshold.
Since the ACT image is generated from the T1w image, which very often has more brainstem in its FoV, the streamlines will be interpreted by ACT to have terminated in white matter, and will therefore be excluded .
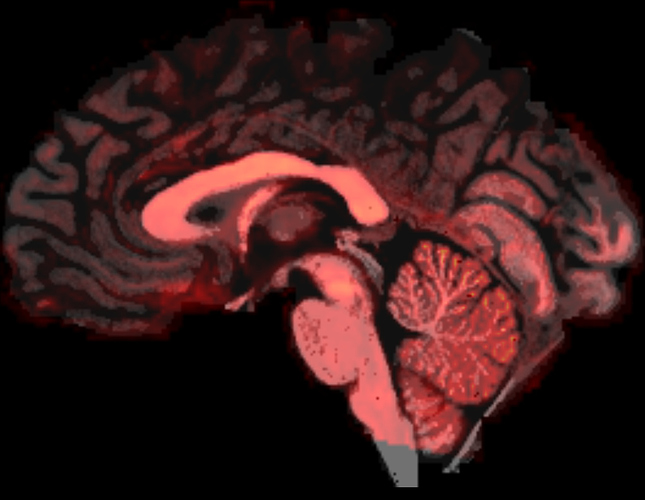
Here is an image of the extent of the DWI image (red overlay) versus the 5TT image (grey):
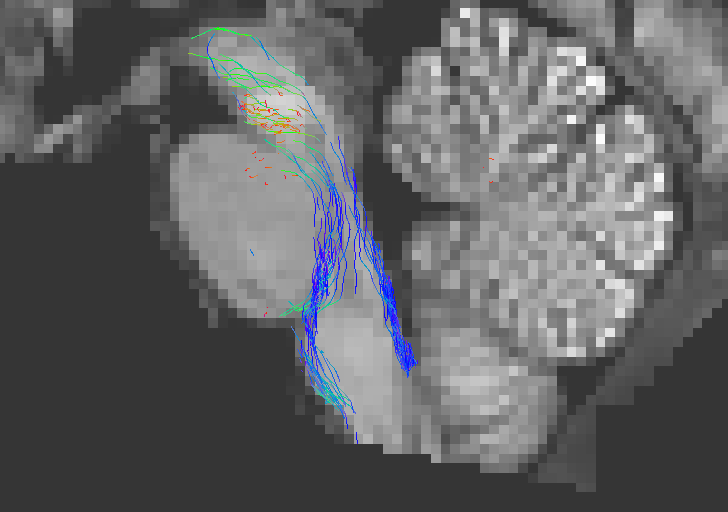
Running tckgen with ACT on these data using this command:
tckgen WM_FODs.mif -seed_sphere 0,-33,-47,10 -seed_dir 0,0,1 -select 1k -act 5TT.mif test_act.tck
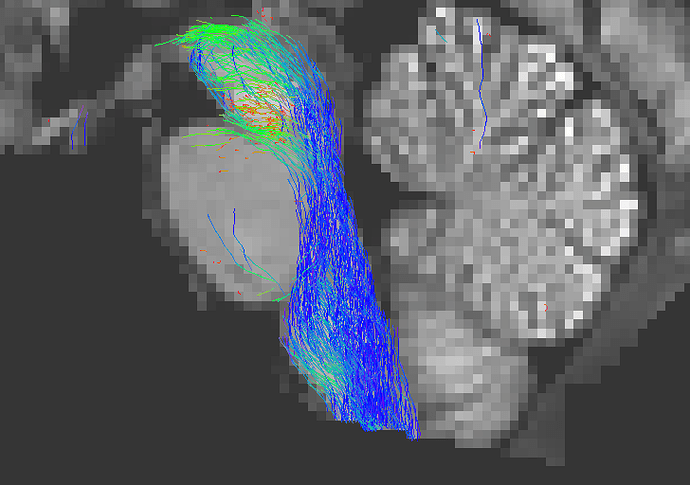
Causes this problem:
As you can see above, the streamlines that terminate on leaving the brainstem are excluded from the .tck file. Note that the streamlines that are visible here are those that terminate in areas of the brainstem that are labeled (potentially mislabeled) as grey matter.
Workarounds
The possible solutions to the problem are various (and rather simple) but none are necessarily “ideal”. The issue should be fixed in the upcoming version of MRtrix3 , but until then, these workarounds could be used.
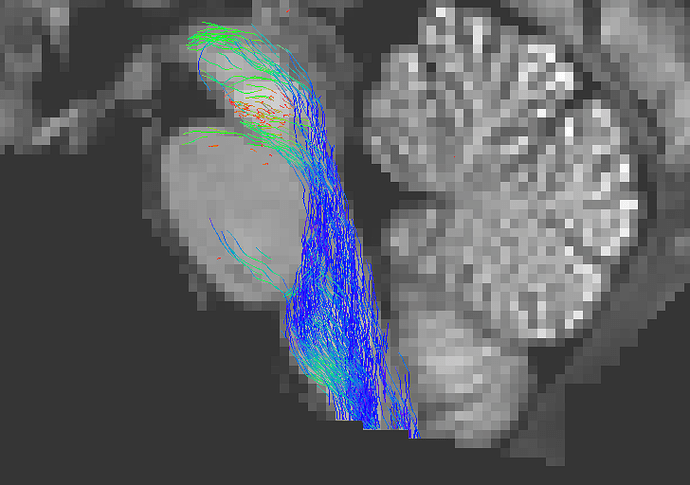
a) Using the -mask option in tckgen
The HCP provides a file called nodif_brain_mask . This handy volume contains a mask of all the voxels that have valid diffusion data. The tckgen command can therefore be run providing this image using the -mask option:
tckgen WM_FODs.mif -seed_sphere 0,-33,-47,10 -seed_dir 0,0,1 -select 1k -act 5TT.mif test_act_mask.tck -mask nodif_brain_mask.nii.gz
As you can see this solves the greater part of the problem. This is possible because providing a 5TT image in order to perform ACT, and providing one or more mask images to constrain where streamlines are permitted to exist, are in fact not mutually exclusive .
Note that premasking the ACT image with the diffusion mask has the same effect.
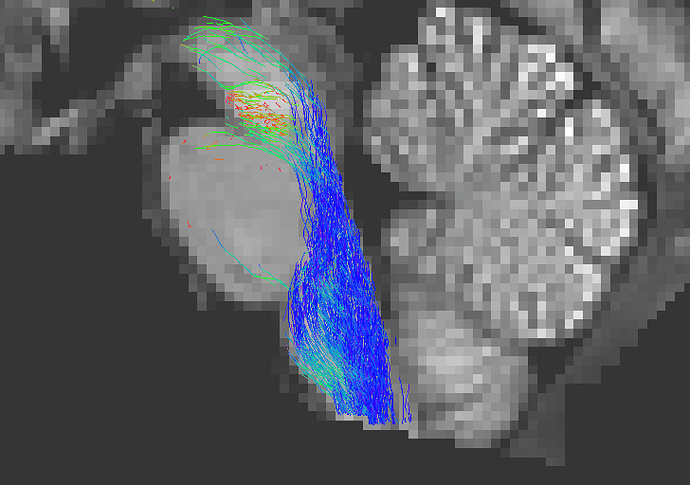
b) Replacing the zeros in the WM_FODs with NaNs
Using the following command, you can explicitly set any voxel with a value of exactly zero to be considered as “not a number” (“NaN”):
mrconvert WM_FODs.mif -coord 3 0 - | mrcalc - 0 -neq WM_FODs.mif nan -if WM_FODs_nan.mif
This results in FODs outside of the brain volume containing “illegal” values (rather than zero values), and is also a potential fix to the problem, since streamlines traversing this region will be terminated due to “absence of valid FOD data” (rather than FOD amplitudes becoming progressively smaller):
tckgen WM_FODs_nan.mif -seed_sphere 0,-33,-47,10 -seed_dir 0,0,1 -select 1k -act 5TT.mif test_act_nan.tck
c) Not using ACT
Finally, although this is not the recommended approach, not using ACT in the first place will also work!
tckgen WM_FODs.mif -seed_sphere 0,-33,-47,10 -seed_dir 0,0,1 -select 1k test_noact.tck
Due to the nature of the HCP data, this error occurs in a high proportion of subjects, and will of course extend to any data that share the same properties (ie brainstem not at the bottom of the FOV).