Dear experts,
my objective is to do tractography in mouse brain using your software but I am able to detect tracts only located the subcortical gray matter:
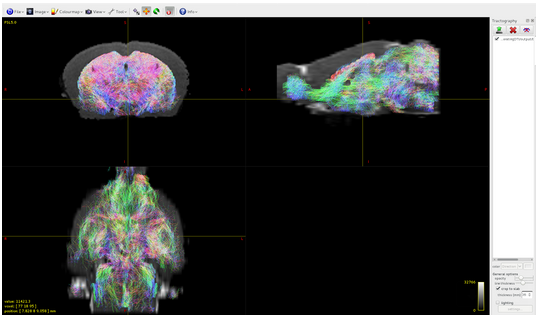
I have the impression that the problem is in the 5TT file that I built by myself, however, since I am not an expert, I will summarize here what I have done counting on your help.
I follow more or less the step described in the HCP tutorial.
DIFFUSION IMAGE PREPROCESSING
mrconvert data.nii.gz DWI.mif -fslgrad bvecs bvals -datatype float32 -stride 0,0,0,1
dwi.mif generated:
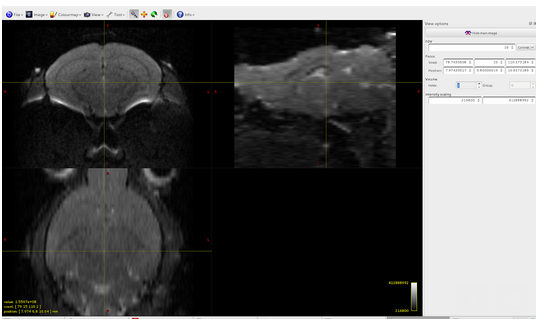
dwi2mask:
dwi2response -nthreads 8 DWI.mif response.txt -mask mask.mif
response.txt: 155223424 -39325424 9820988 -1983875.25
dwi2fod -nthreads 8 DWI.mif response.txt csd.mif -mask mask.mif
csd.mif volume 0:
csd.mif, volume1:
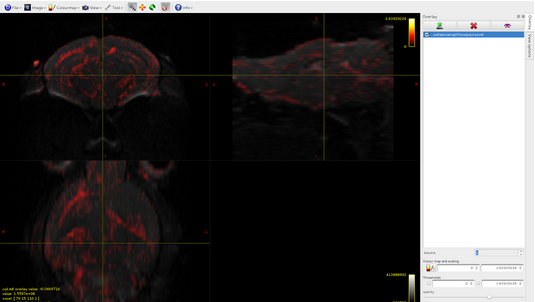
The csd file is composed of 28 volumes, and if I well understand, this means that I have 28 different spherical harmonic components, is it reasonable?
STRUCTURAL IMAGE PROCESSING:
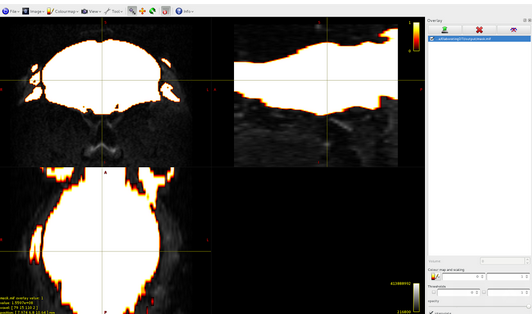
I built by myself the 5TT file (that you can find attached here) and I registered it in the diffusion space (also if I think that it is not strictly necessary). Respecting the instruction that I found in your documentation, the 5TT file is a 4D 32 bit floating image, where the dimension of the 4th axis is five: here there are the images of the overlap between the dwi.mif file and the 5 volumes of the 5TT file that I built:
Volume0: cortical gray matter
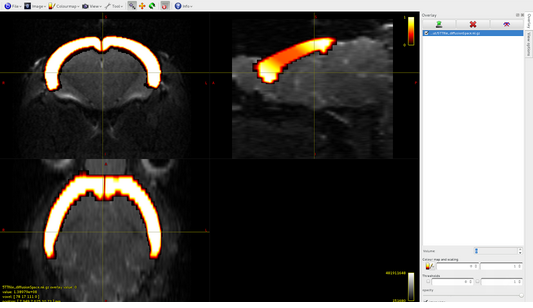
Volume1: subcortical gray matter
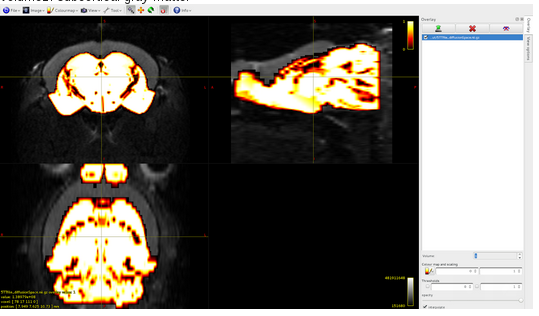
Volume2: white matter
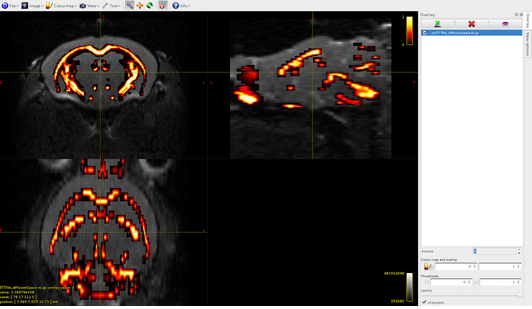
Volume3: CSF
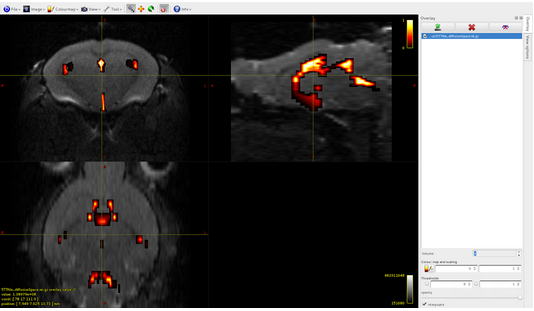
Volume4: pathological tissue
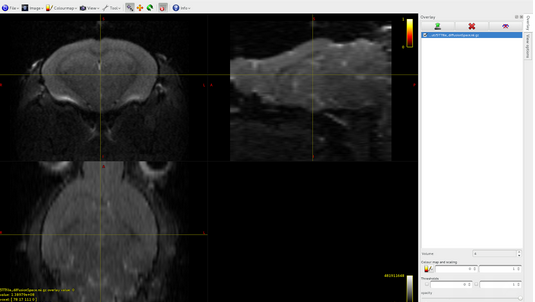
mrconvert 5TTfile_diffusionSpace.nii.gz 5TTfile.mif5tt2gmwmi 5TTfile.mif gmwmi.mif
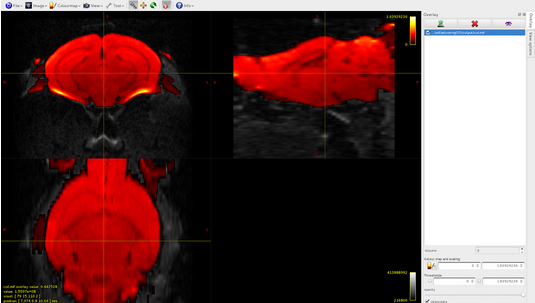
Here it is the overlap between dwi and gmwmi
CONNECTOME GENERATION:
tckgen -nthreads 8 -num 500000 -seed_gmwmi gmwmi.mif -act
5TTfile_diffusionSpace.mif -mask mask.mif -unidirectional -maxlength 100000 -step 0.05 csd.mif brain.tck
(I know that I used big values for -num, -maxlength and -step, but was to exclude that was that one the problem)
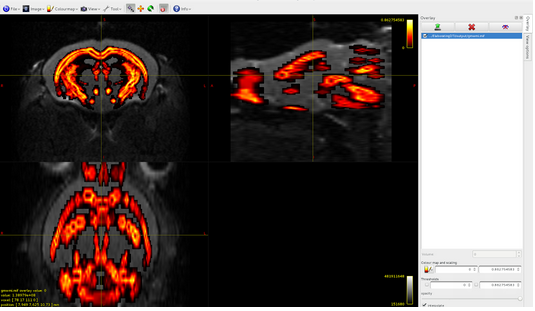
tcksift brain.tck csd.mif brain-sift.tck -act 5TTfile_diffusionSpace.mif
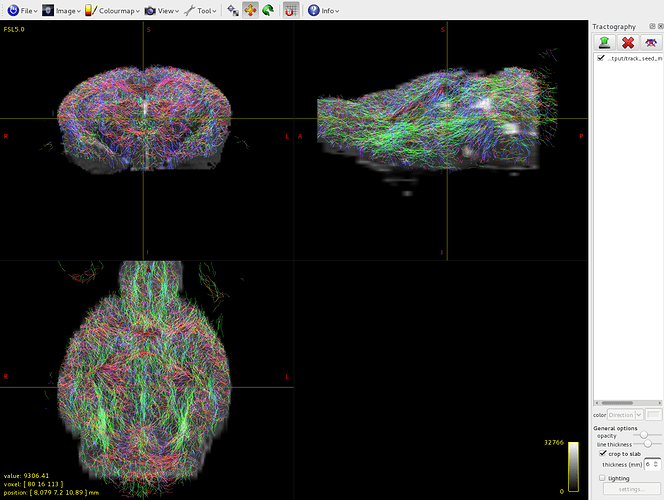
Here it is the overlap between brain-sift.tck and dwi:
It seems that the algorithm detects tracts only in volume 1 of the 5tt file, where am I wrong?
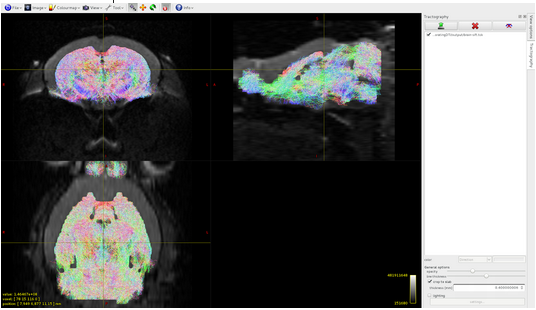
The problem is solved when I use as seed image for tckgen the dwi mask (as described in your documentation in the tutorial Basic DWI processing):
Please find here https://www.dropbox.com/sh/knxoj5wnfoekpvb/AADP8E0nQaMZESoi4UJWzGZqa?dl=0 the 5TT file that I used.
Thanks in advance for your time,
Francesca
 now I will need time to do the racetrack step by step
now I will need time to do the racetrack step by step