Hello everyone,
I tried to combine segmented lesion mask with 5tt image in order to perform global tractography.
I used this command
*5ttedit –path segmentedlesion.mif 5ttimage.mif 5tt_lesion.mif
But I encountered the problem with mismatch of two images dimensions. The error message was:
Image segmenedlesion_mask.mif does not match 5TT image dimensions
Then, I checked the size of images which is displayed below, and resized size of lesion file using:
*mrresize –size 142,156,142 input –interp linear output
mrinfo 5tt_nocoreg.mif
Image: “5tt_nocoreg.mif”
Dimensions: 142 x 156 x 142 x 5
Voxel size: 1 x 1 x 1 x ?
Data strides: [ 2 3 4 1 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 0.9921 0.03527 -0.1206 -67.01
-0.01916 0.991 0.1322 -68.2
0.1242 -0.1288 0.9839 -58.05
mrinfo segmentedlesion.nii
Image: “segmentedlesion.nii”
Dimensions: 192 x 456 x 512
Voxel size: 1 x 0.488281 x 0.488281
Data strides: [ 1 2 3 ]
Format: NIfTI-1.1
Data type: unsigned 8 bit integer
Intensity scaling: offset = 0, multiplier = 1
Transform: 0.9921 0.03527 -0.1206 -81.84
-0.01916 0.991 0.1322 -92.29
0.1242 -0.1288 0.9839 -136.5
comments: TE=3.9e+02;Time=92512.870;phase=1
mrinfo segmentedlesion_mask_resize_linear.mif
Image: “segmentedlesion_mask_resize_linear.mif”
Dimensions: 142 x 156 x 142
Voxel size: 1.35211 x 1.42728 x 1.76056
Data strides: [ 1 2 3 ]
Format: MRtrix
Data type: 32 bit float (little endian)
Intensity scaling: offset = 0, multiplier = 1
Transform: 0.9921 0.03527 -0.1206 -81.73
-0.01916 0.991 0.1322 -91.75
0.1242 -0.1288 0.9839 -135.9
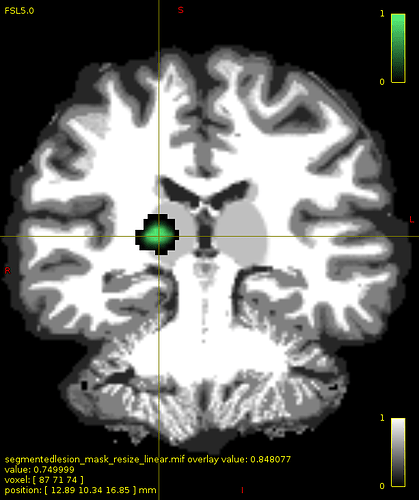
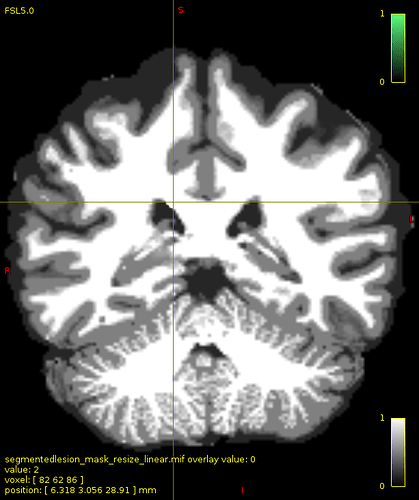
Then,I ran 5ttedit to merge lesion with 5tt image. In order to check if the lesion has been combined correctly, I ran 5tt2vis. Surprisingly, the position of lesion was somewhere else. As you can see in image, the green region is the lesion overlaid on 5tt image and in second image the crosshair is on the lesion incorporated in 5ttimage with value of 2 (default value for pathological tissue in 5tt2vis). The question is, why does 5ttedit change the image position while it has the same dimension as 5ttimage.
!One more question, mrresize changed voxel size in addition to dimensions, does it make any kind of problems ?