Hello all,
I wonder whether anyone here has experience using 5ttgen or any other script for reasonable tissue segmentation of T1w or T2w of infants?
I need the segmentation results for Anatomically Constrained Tractography.
Thank you,
Mahmoud
Hello all,
I wonder whether anyone here has experience using 5ttgen or any other script for reasonable tissue segmentation of T1w or T2w of infants?
I need the segmentation results for Anatomically Constrained Tractography.
Thank you,
Mahmoud
That would depend heavily on how old the infants are. If they’re older than 2 or so, there’s a good chance the default approaches in 5ttgen might work out of the box. But neonates definitely need to be handled differently, due to the very different contrast in the images. In this age range, dedicated packages like DrawEM work much better – but it’ll mean you’ll need to create your 5TT images manually, sidestepping the 5ttgen script altogether – shouldn’t be too difficult once you’ve figured out what’s expected.
Thanks for your response. Yes, the challenge is that the infants are younger than 6-months.
In this age range, dedicated packages like DrawEM work much better – but it’ll mean you’ll need to create your 5TT images manually …
Or you could write a 5ttgen algorithm based on such software and contribute it to MRtrix3 ![]()
@rsmith or @jdtournier: Perhaps the below could be made an issue on https://github.com/mrtrix3???
Hi @zeydabadi,
I actually created such a modification to 5ttgen for a previous version of mrtrix3 to take DrawEM segmentations into ACT. Now, I am updating it to work with the latest implementation of mrtrix3, in which the 5ttgen script has changed. To modify the 5ttgen current script see, as @jdtournier pointed out, 5ttgen documentation.
Here is an algorithm I have created to use DrawEM tissue labels in 5ttgen.
$ cat mrtrix3/lib/mrtrix3/_5ttgen/drawem_tissue_labels.py
def initialise(base_parser, subparsers): #pylint: disable=unused-variable
parser = subparsers.add_parser('drawem_tissue_labels', author='Finn Lennartsson (finn.lennartsson@gmail.com)', synopsis='Generate the 5TT image based on a DrawEM ALBERTs neonatal tissue label parcellation image', parents=[base_parser])
parser.add_argument('input', help='The input the DrawEM neonatal tissue label parcellation image)')
parser.add_argument('output', help='The output 5TT image')
options = parser.add_argument_group('Options specific to the \'drawem_tissue_labels\' algorithm')
options.add_argument('-lut', help='Manually provide path to the lookup table on which the input parcellation image is based (e.g. $DRAWEMDIR/label_names_ACT/tissue_labels.txt)')
def checkOutputPaths(): #pylint: disable=unused-variable
pass
def getInputs(): #pylint: disable=unused-variable
import shutil
from mrtrix3 import app, path, run
run.command('mrconvert ' + path.fromUser(app.args.input, True) + ' ' + path.toTemp('input.mif', True))
if app.args.lut:
run.function(shutil.copyfile, path.fromUser(app.args.lut, False), path.toTemp('LUT.txt', False))
def execute(): #pylint: disable=unused-variable
import os.path #pylint: disable=unused-variable
from mrtrix3 import app, path, run
lut_input_path = 'LUT.txt'
if not os.path.exists('LUT.txt'):
drawem_home = os.environ.get('DRAWEMDIR', '')
if not drawem_home:
app.error('Environment variable DRAWEMDIR is not set; please run appropriate configuration script, set this variable manually, or provide script with path to appropriate DrawEM LUT file using -lut option')
lut_input_path = os.path.join(drawem_home, 'label_names_ACT','tissue_labels.txt')
if not os.path.isfile(lut_input_path):
app.error('Could not find DrawEM lookup table file (expected location: ' + lut_input_path + '), and none provided using -lut')
if app.args.sgm_amyg_hipp:
lut_output_file_name = 'DrawEM_tissue_labels2ACT_sgm_amyg_hipp.txt'
else:
lut_output_file_name = 'DrawEM_tissue_labels2ACT.txt'
#lut_output_path = os.path.join(os.path.dirname(os.path.abspath(sys.argv[0])), 'data', lut_output_file_name);
lut_output_path = os.path.join(path.sharedDataPath(), path.scriptSubDirName(), lut_output_file_name)
if not os.path.isfile(lut_output_path):
app.error('Could not find lookup table file for converting DrawEM parcellation output to tissues (expected location: ' + lut_output_path + ')')
# Initial conversion from DrawEM tissue label parcellation to five principal tissue types
run.command('labelconvert input.mif ' + lut_input_path + ' ' + lut_output_path + ' indices.mif')
# Use mrcrop to reduce file size
if app.args.nocrop:
image = 'indices.mif'
else:
image = 'indices_cropped.mif'
run.command('mrthreshold indices.mif - -abs 0.5 | mrcrop indices.mif ' + image + ' -mask -')
# Convert into the 5TT format for ACT
run.command('mrcalc ' + image + ' 1 -eq cgm.mif')
run.command('mrcalc ' + image + ' 2 -eq sgm.mif')
run.command('mrcalc ' + image + ' 3 -eq wm.mif')
run.command('mrcalc ' + image + ' 4 -eq csf.mif')
run.command('mrcalc ' + image + ' 5 -eq path.mif')
run.command('mrcat cgm.mif sgm.mif wm.mif csf.mif path.mif - -axis 3 | mrconvert - result.mif -datatype float32')
As the input LUT, I use a modification of the DrawEM tissue label csv-file where a basically just put in hyphens to make the to make the it work with labelconvert.
$ cat tissue_labels.txt
1 CSF
2 Cortical-gray-matter
3 White-matter
4 Background
5 Ventricles
6 Cerebellum
7 Deep-Gray-Matter
8 Brainstem
9 Hippocampi-and-Amygdala
This file (tissue_labels.txt) lives my $DRAWEMDIR/label_names_ACT, as seen in the py-file, but I suggest you modify the file and put the input LUT in the mrtrix3/share/mrtrix3/_5ttgen/, where the output LUT lives.
These are
$ cat mrtrix3/share/mrtrix3/_5ttgen/DrawEM_tissue_labels2ACT.txt
4 CSF
1 Cortical-gray-matter
3 White-matter
0 Background
4 Ventricles
1 Cerebellum
2 Deep-Gray-Matter
3 Brainstem
1 Hippocampi-and-Amygdala
and
$ cat mrtrix3/share/mrtrix3/_5ttgen/DrawEM_tissue_labels2ACT_sgm_amyg_hipp.txt
4 CSF
1 Cortical-gray-matter
3 White-matter
0 Background
4 Ventricles
1 Cerebellum
2 Deep-Gray-Matter
3 Brainstem
2 Hippocampi-and-Amygdala
I have also created corresponding LUTs for the all_labels.csv
$ cat mrtrix3/share/mrtrix3/_5ttgen/DrawEM_all_labels.txt
1 Hippocampus-left
2 Hippocampus-right
3 Amygdala-left
4 Amygdala-right
5 Anterior-temporal-lobe,-medial-part-left-GM
6 Anterior-temporal-lobe,-medial-part-right-GM
7 Anterior-temporal-lobe,-lateral-part-left-GM
8 Anterior-temporal-lobe,-lateral-part-right-GM
9 Gyri-parahippocampalis-et-ambiens-anterior-part-left-GM
10 Gyri-parahippocampalis-et-ambiens-anterior-part-right-GM
11 Superior-temporal-gyrus,-middle-part-left-GM
12 Superior-temporal-gyrus,-middle-part-right-GM
13 Medial-and-inferior-temporal-gyri-anterior-part-left-GM
14 Medial-and-inferior-temporal-gyri-anterior-part-right-GM
15 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-GM
16 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-GM
17 Cerebellum-left
18 Cerebellum-right
19 Brainstem,-spans-the-midline
20 Insula-right-GM
21 Insula-left-GM
22 Occipital-lobe-right-GM
23 Occipital-lobe-left-GM
24 Gyri-parahippocampalis-et-ambiens-posterior-part-right-GM
25 Gyri-parahippocampalis-et-ambiens-posterior-part-left-GM
26 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-GM
27 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-GM
28 Medial-and-inferior-temporal-gyri-posterior-part-right-GM
29 Medial-and-inferior-temporal-gyri-posterior-part-left-GM
30 Superior-temporal-gyrus,-posterior-part-right-GM
31 Superior-temporal-gyrus,-posterior-part-left-GM
32 Cingulate-gyrus,-anterior-part-right-GM
33 Cingulate-gyrus,-anterior-part-left-GM
34 Cingulate-gyrus,-posterior-part-right-GM
35 Cingulate-gyrus,-posterior-part-left-GM
36 Frontal-lobe-right-GM
37 Frontal-lobe-left-GM
38 Parietal-lobe-right-GM
39 Parietal-lobe-left-GM
40 Caudate-nucleus-right
41 Caudate-nucleus-left
42 Thalamus-right,-high-intensity-part-in-T2
43 Thalamus-left,-high-intensity-part-in-T2
44 Subthalamic-nucleus-right
45 Subthalamic-nucleus-left-
46 Lentiform-Nucleus-right
47 Lentiform-Nucleus-left
48 Corpus-Callosum
49 Lateral-Ventricle-left
50 Lateral-Ventricle-right
51 Anterior-temporal-lobe,-medial-part-left-WM
52 Anterior-temporal-lobe,-medial-part-right-WM
53 Anterior-temporal-lobe,-lateral-part-left-WM
54 Anterior-temporal-lobe,-lateral-part-right-WM
55 Gyri-parahippocampalis-et-ambiens-anterior-part-left-WM
56 Gyri-parahippocampalis-et-ambiens-anterior-part-right-WM
57 Superior-temporal-gyrus,-middle-part-left-WM
58 Superior-temporal-gyrus,-middle-part-right-WM
59 Medial-and-inferior-temporal-gyri-anterior-part-left-WM
60 Medial-and-inferior-temporal-gyri-anterior-part-right-WM
61 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-WM
62 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-WM
63 Insula-right-WM
64 Insula-left-WM
65 Occipital-lobe-right-WM
66 Occipital-lobe-left-WM
67 Gyri-parahippocampalis-et-ambiens-posterior-part-right-WM
68 Gyri-parahippocampalis-et-ambiens-posterior-part-left-WM
69 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-WM
70 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-WM
71 Medial-and-inferior-temporal-gyri-posterior-part-right-WM
72 Medial-and-inferior-temporal-gyri-posterior-part-left-WM
73 Superior-temporal-gyrus,-posterior-part-right-WM
74 Superior-temporal-gyrus,-posterior-part-left-WM
75 Cingulate-gyrus,-anterior-part-right-WM
76 Cingulate-gyrus,-anterior-part-left-WM
77 Cingulate-gyrus,-posterior-part-right-WM
78 Cingulate-gyrus,-posterior-part-left-WM
79 Frontal-lobe-right-WM
80 Frontal-lobe-left-WM
81 Parietal-lobe-right-WM
82 Parietal-lobe-left-WM
83 CSF
84 Extra-cranial-background
85 Intra-cranial-background
86 Thalamus-right,-low-intensity-part-in-T2
87 Thalamus-left,-low-intensity-part-in-T2
and corresponding 2ACT-maps
$ cat mrtrix3/share/mrtrix3/_5ttgen/DrawEM_all_labels2ACT.txt
4 CSF
1 Anterior-temporal-lobe,-medial-part-left-GM
1 Anterior-temporal-lobe,-medial-part-right-GM
1 Anterior-temporal-lobe,-lateral-part-left-GM
1 Anterior-temporal-lobe,-lateral-part-right-GM
1 Gyri-parahippocampalis-et-ambiens-anterior-part-left-GM
1 Gyri-parahippocampalis-et-ambiens-anterior-part-right-GM
1 Superior-temporal-gyrus,-middle-part-left-GM
1 Superior-temporal-gyrus,-middle-part-right-GM
1 Medial-and-inferior-temporal-gyri-anterior-part-left-GM
1 Medial-and-inferior-temporal-gyri-anterior-part-right-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-GM
1 Insula-right-GM
1 Insula-left-GM
1 Occipital-lobe-right-GM
1 Occipital-lobe-left-GM
1 Gyri-parahippocampalis-et-ambiens-posterior-part-right-GM
1 Gyri-parahippocampalis-et-ambiens-posterior-part-left-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-GM
1 Medial-and-inferior-temporal-gyri-posterior-part-right-GM
1 Medial-and-inferior-temporal-gyri-posterior-part-left-GM
1 Superior-temporal-gyrus,-posterior-part-right-GM
1 Superior-temporal-gyrus,-posterior-part-left-GM
1 Cingulate-gyrus,-anterior-part-right-GM
1 Cingulate-gyrus,-anterior-part-left-GM
1 Cingulate-gyrus,-posterior-part-right-GM
1 Cingulate-gyrus,-posterior-part-left-GM
1 Frontal-lobe-right-GM
1 Frontal-lobe-left-GM
1 Parietal-lobe-right-GM
1 Parietal-lobe-left-GM
3 Corpus-Callosum
3 Anterior-temporal-lobe,-medial-part-left-WM
3 Anterior-temporal-lobe,-medial-part-right-WM
3 Anterior-temporal-lobe,-lateral-part-left-WM
3 Anterior-temporal-lobe,-lateral-part-right-WM
3 Gyri-parahippocampalis-et-ambiens-anterior-part-left-WM
3 Gyri-parahippocampalis-et-ambiens-anterior-part-right-WM
3 Superior-temporal-gyrus,-middle-part-left-WM
3 Superior-temporal-gyrus,-middle-part-right-WM
3 Medial-and-inferior-temporal-gyri-anterior-part-left-WM
3 Medial-and-inferior-temporal-gyri-anterior-part-right-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-WM
3 Insula-right-WM
3 Insula-left-WM
3 Occipital-lobe-right-WM
3 Occipital-lobe-left-WM
3 Gyri-parahippocampalis-et-ambiens-posterior-part-right-WM
3 Gyri-parahippocampalis-et-ambiens-posterior-part-left-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-WM
3 Medial-and-inferior-temporal-gyri-posterior-part-right-WM
3 Medial-and-inferior-temporal-gyri-posterior-part-left-WM
3 Superior-temporal-gyrus,-posterior-part-right-WM
3 Superior-temporal-gyrus,-posterior-part-left-WM
3 Cingulate-gyrus,-anterior-part-right-WM
3 Cingulate-gyrus,-anterior-part-left-WM
3 Cingulate-gyrus,-posterior-part-right-WM
3 Cingulate-gyrus,-posterior-part-left-WM
3 Frontal-lobe-right-WM
3 Frontal-lobe-left-WM
3 Parietal-lobe-right-WM
3 Parietal-lobe-left-WM
0 Extra-cranial-background
4 Lateral-Ventricle-left
4 Lateral-Ventricle-right
1 Cerebellum-left
1 Cerebellum-right
2 Caudate-nucleus-right
2 Caudate-nucleus-left
2 Thalamus-right,-high-intensity-part-in-T2
2 Thalamus-left,-high-intensity-part-in-T2
2 Subthalamic-nucleus-right
2 Subthalamic-nucleus-left-
2 Lentiform-Nucleus-right
2 Lentiform-Nucleus-left
2 Intra-cranial-background
2 Thalamus-right,-low-intensity-part-in-T2
2 Thalamus-left,-low-intensity-part-in-T2
3 Brainstem,-spans-the-midline
1 Hippocampus-left
1 Hippocampus-right
1 Amygdala-left
1 Amygdala-right
and
$ cat mrtrix3/share/mrtrix3/_5ttgen/DrawEM_all_labels2ACT_sgm_amyg_hipp.txt
ACT-value All-labels
4 CSF
1 Anterior-temporal-lobe,-medial-part-left-GM
1 Anterior-temporal-lobe,-medial-part-right-GM
1 Anterior-temporal-lobe,-lateral-part-left-GM
1 Anterior-temporal-lobe,-lateral-part-right-GM
1 Gyri-parahippocampalis-et-ambiens-anterior-part-left-GM
1 Gyri-parahippocampalis-et-ambiens-anterior-part-right-GM
1 Superior-temporal-gyrus,-middle-part-left-GM
1 Superior-temporal-gyrus,-middle-part-right-GM
1 Medial-and-inferior-temporal-gyri-anterior-part-left-GM
1 Medial-and-inferior-temporal-gyri-anterior-part-right-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-GM
1 Insula-right-GM
1 Insula-left-GM
1 Occipital-lobe-right-GM
1 Occipital-lobe-left-GM
1 Gyri-parahippocampalis-et-ambiens-posterior-part-right-GM
1 Gyri-parahippocampalis-et-ambiens-posterior-part-left-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-GM
1 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-GM
1 Medial-and-inferior-temporal-gyri-posterior-part-right-GM
1 Medial-and-inferior-temporal-gyri-posterior-part-left-GM
1 Superior-temporal-gyrus,-posterior-part-right-GM
1 Superior-temporal-gyrus,-posterior-part-left-GM
1 Cingulate-gyrus,-anterior-part-right-GM
1 Cingulate-gyrus,-anterior-part-left-GM
1 Cingulate-gyrus,-posterior-part-right-GM
1 Cingulate-gyrus,-posterior-part-left-GM
1 Frontal-lobe-right-GM
1 Frontal-lobe-left-GM
1 Parietal-lobe-right-GM
1 Parietal-lobe-left-GM
3 Corpus-Callosum
3 Anterior-temporal-lobe,-medial-part-left-WM
3 Anterior-temporal-lobe,-medial-part-right-WM
3 Anterior-temporal-lobe,-lateral-part-left-WM
3 Anterior-temporal-lobe,-lateral-part-right-WM
3 Gyri-parahippocampalis-et-ambiens-anterior-part-left-WM
3 Gyri-parahippocampalis-et-ambiens-anterior-part-right-WM
3 Superior-temporal-gyrus,-middle-part-left-WM
3 Superior-temporal-gyrus,-middle-part-right-WM
3 Medial-and-inferior-temporal-gyri-anterior-part-left-WM
3 Medial-and-inferior-temporal-gyri-anterior-part-right-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-left-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-anterior-part-right-WM
3 Insula-right-WM
3 Insula-left-WM
3 Occipital-lobe-right-WM
3 Occipital-lobe-left-WM
3 Gyri-parahippocampalis-et-ambiens-posterior-part-right-WM
3 Gyri-parahippocampalis-et-ambiens-posterior-part-left-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-right-WM
3 Lateral-occipitotemporal-gyrus,-gyrus-fusiformis-posterior-part-left-WM
3 Medial-and-inferior-temporal-gyri-posterior-part-right-WM
3 Medial-and-inferior-temporal-gyri-posterior-part-left-WM
3 Superior-temporal-gyrus,-posterior-part-right-WM
3 Superior-temporal-gyrus,-posterior-part-left-WM
3 Cingulate-gyrus,-anterior-part-right-WM
3 Cingulate-gyrus,-anterior-part-left-WM
3 Cingulate-gyrus,-posterior-part-right-WM
3 Cingulate-gyrus,-posterior-part-left-WM
3 Frontal-lobe-right-WM
3 Frontal-lobe-left-WM
3 Parietal-lobe-right-WM
3 Parietal-lobe-left-WM
0 Extra-cranial-background
4 Lateral-Ventricle-left
4 Lateral-Ventricle-right
1 Cerebellum-left
1 Cerebellum-right
2 Caudate-nucleus-right
2 Caudate-nucleus-left
2 Thalamus-right,-high-intensity-part-in-T2
2 Thalamus-left,-high-intensity-part-in-T2
2 Subthalamic-nucleus-right
2 Subthalamic-nucleus-left-
2 Lentiform-Nucleus-right
2 Lentiform-Nucleus-left
2 Intra-cranial-background
2 Thalamus-right,-low-intensity-part-in-T2
2 Thalamus-left,-low-intensity-part-in-T2
3 Brainstem,-spans-the-midline
2 Hippocampus-left
2 Hippocampus-right
2 Amygdala-left
2 Amygdala-right
Hi @Finn,
Very interesting work, I’m very interested on this. Could you show an example of how it looks? It will be fantastic having everything implemented together in MRtrix.
I use another approach to create the 5TT file for ACT, making use of TPM obtained from DrawEM and the M-CRIB atlas, but I’m not familiar with python, so I do everything in bash.
Best regards,
Manuel
Thank you so much for sharing!
I’ve also learned that maybe infantFS is better option for my cohort’s age range.
Has anyone been able to try it?
Hi @mblesac,
My interest has been to study the functional and structural connectivity in neonates.
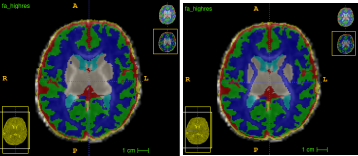
The main objective for using DrawEM has been to use it to create the 5TT file for ACT. For this purpose, I focused on the tissue label output from DrawEM. I had to manually edit this as the central non-myelinated WM is misclassified on T2w (left pic). However, a FA depicts it well, so I used that as the template while editing (right pic).
I then use my modified “5ttgen drawem_tissue_labels”-command to take this into 5tt:
5ttgen drawem_tissue_labels DrawEM_tissue_label_edited.nii.gz 5tt.mif
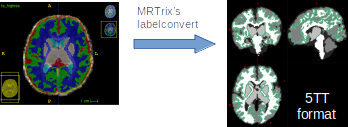
(Under the 5ttgen-hood it is a labelconvert-command.)
My work is published here, but I can send you a copy if you cannot access the pdf.
So the painstaking step is the manual editing, but it doesn’t take too long. The improved “tissue label” maps can also be used to refine the “all labels” or the “labels”-map. I used ITK-SNAP for editing (here are LUTs).
On another note. Perhaps the FA could augment the segmentation?!?
I also find the current parcellation scheme, with its 50 brain regions, too limited (e.g. each temporal lobe contains 12 regions compared to one single region for the whole frontal lobe). Have you any plans on making your work available? That would be fantastic!
Hope that is to some help!
Finn
@rsmith or @jdtournier: Perhaps the below could be made an issue on MRtrix3 · GitHub???
Absolutely! Best approach is to make a fork of the MRtrix3 repository using your own GitHub account, commit the changes into a new branch, and then create a Pull Request to bring the changes from your fork into the MRtrix3 repository. That way git will properly attribute to your account the relevant changes.
For non-bugfixes, we want changes to be merged into the dev code branch. However the dev branch currently contains some decent changes to the Python API that will be incorporated in the next update. I can walk you through the changes that need to be made to the source code on GitHub (or I might write a documentation page describing how to port code to the new API, and you can be my guinea pig to make sure the instructions are adequate ![]() ).
).
Hi Rob,
Sorry for late reply. Has been off on holidays and then back to clinical work with on-call night shift week.
Great suggestion, but I don’t really have the knowledge to manage what you suggest at the moment, unfortunately. So you better find a more appropriate guinea pig. Sorry for that. Will have a closer look again in November when I am back doing research. Perhaps I can step up…
/Finn
Hi
I need to perform multi shell multi tissue CSD on neonatal cohort acquired with HARDI protocol.
However, as structural images I just have 3dT1 as volumetric scans, not T2. All pipelines for segmentation (needed to construct 5ttgen image) resort to T2 images. Does anyone know a method which uses T1?
thanks
Rosella