Dear MRtrix experts,
Hi! I’ve encountered an error while running the 5ttgen command on my data.
I’ve ran this line of commands:
5ttgen fsl anat.mif 5tt_nocoreg.mif
which, in the log file, contains the following:
mrconvert /Users/server/Desktop/sub-032302/ses-01/dwi/anat.mif /Users/server/Desktop/sub-032302/ses-01/dwi/5ttgen-tmp-5Z3S8K/input.mif
mrconvert input.mif T1.nii -strides -1,+2,+3
maskfilter /usr/local/fsl/data/standard/MNI152_T1_1mm_brain_mask_dil.nii.gz dilate mni_mask.nii -npass 4
standard_space_roi T1.nii T1_preBET.nii.gz -maskMASK mni_mask.nii -roiFOV
bet T1_preBET.nii.gz T1_BET.nii.gz -f 0.15 -R
fast T1_BET.nii.gz
run_first_all -m none -s L_Accu,R_Accu,L_Caud,R_Caud,L_Pall,R_Pall,L_Puta,R_Puta,L_Thal,R_Thal -i T1.nii -o first
and got an error message.
[ERROR] FSL FIRST has failed; 0 of 10 structures were segmented successfully (check /Users/server/Desktop/sub-032302/ses-01/dwi/5ttgen-tmp-RA22G8/first.logs)
We’ve tried the -debug option to find out the problem, but we still got the exact same message, nothing else.
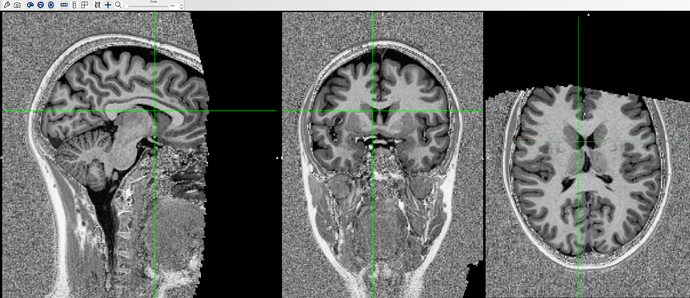
We checked on our T1.nii image and confirmed that it is okay, but then we found out that our T1_preBET image was completely cropped out like the following:
so we are suspecting that something went wrong while executing the standard_space_roi command, possibly aligning the mni mask and the T1 image.
We’ve also checked our mni_mask.nii file to be sure, and it was okay too.
What might be the problem? If the problem is as we suspect, would there be any way to fix it?
Thank you in advance!