Hi James,
I decided to break this discussion off into its own topic, since it’s no longer specific to the absence of the ACPCDetect command in PATH but is more about the nuances of the output that 5ttgen hsvs generates.
I would say the segmentation is the best I’ve ever seen. I"ll do a more careful look with the segmentations overlayed on the T1 to be sure, but for now I can only say congrats, this is really good, and has all the subcortical white matter nicely partitioned as well, with a very nice job on the fornix–again, best I’ve seen by some distance.
Thanks, but I can only take a fraction of the credit. Ultimately the segmentations are principally from FreeSurfer, with some FSL FIRST in there too; all I’ve done is work on the algorithm for going from surface representations to partial volume fractions, and added some surface-based smoothing to those structures for which FreeSurfer only provides a binary voxel classification.
Question: is it possible, or how might one incorporate the hippocampal subfield and thalamic nuclei segmentation from Freesurfer into the MRtrix 5ttgen output.
This should already be possible, as long as those modules have already been run for that FreeSurfer subject prior to invoking 5ttgen hsvs.
Note however that even if the thalamic nuclei module has been run, 5ttgen hsvs will by default still use FSL FIRST to segment the thalami; it is only if you specify the -thalami nuclei option that the output of that module will be used. I personally found the delineations from FIRST to appear more believable on the data I tried; but I’m happy to be proven wrong.
All looks great except for an area in the center of the brain. I’ve made some screenshot videos of the white matter segmentation overlaid on the T1 and I’ll send them to your florey.au.edu email with an explanation of what I think I see. Looks like there may be a fault in the segmentation around the front end of the fornix.
Thanks for sending those through. I think I understand what you’re indicating, though my anatomical knowledge is limited and therefore my judgement is correspondingly so. If there’s nuance to be understood here a video call would likely reach a mutual understanding faster.
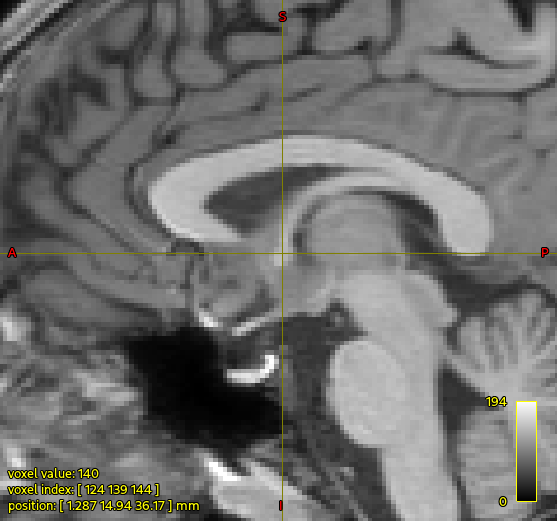
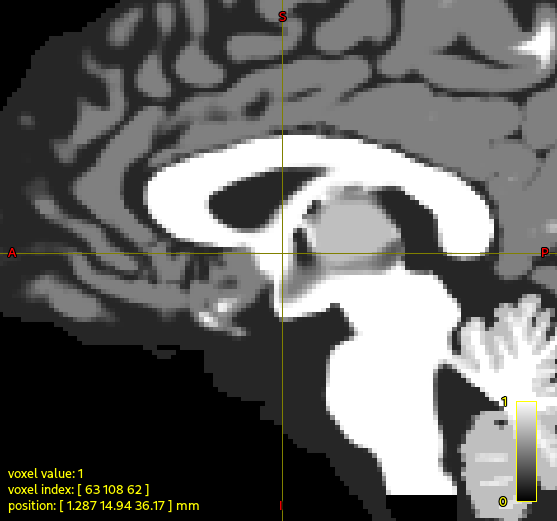
For those playing along at home, here’s the region for which the concern is being expressed:
In the raw T1w image, at the position of the cursor, the fornix looks quite narrow, whereas in the segmented image (here showing the output of 5tt2vis) there’s a kind of big “blob” of white matter.
The trouble here is that FreeSurfer actually labels that whole region as index 2 “Left-Cerebral-White-Matter”, and there’s no other explicit segmentation defined there. So based on the cumulative information provided to the algorithm, that whole region is white matter. In your data the FIRST segmentation of the nucleus accumbens is considerably smaller than that estimated by FreeSurfer, which may be contributing to the anterior portion of that blob. But as far as I can tell there’s nothing going wrong within 5ttgen hsvs, it’s producing what it should given the segmentations with which it has been provided.
If there are other software tools that can provide better segmentations of specific structures, there’s always the possibility of augmenting the 5ttgen algorithm to make use of them. But I’m not personally in the field of T1w image segmentation, I just try to convert what others have done into a form that I can use.
Cheers
Rob