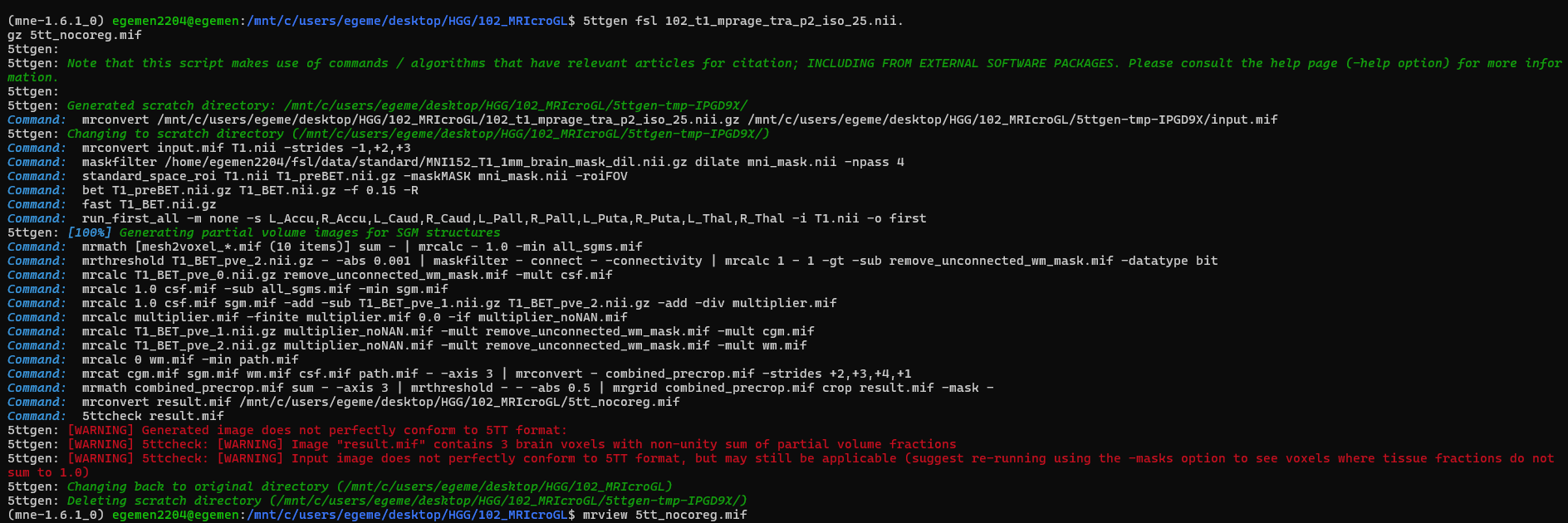
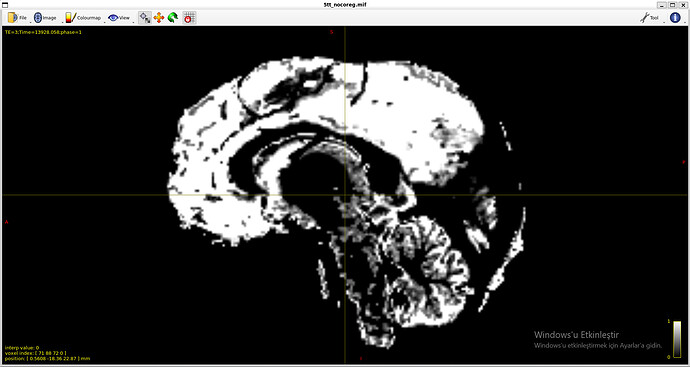
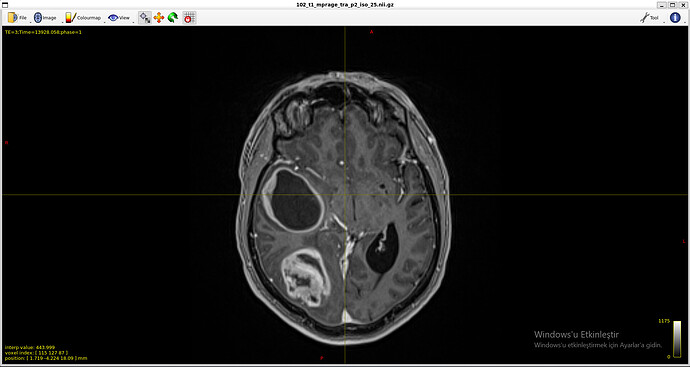
It says in warning message I should use -mask option but there isnt -mask option for 5ttgen.
What should I do in this situation? In my dataset I cant work with any of T1 or Flair images of patients. Either recon-all or 5ttgen is not giving good results
5tt_nocoreg.mif (it is somehow reversed in space)
T1 image
Because of massive tumors, 5ttgen was not working properly.
I used SynthSR algorithm from freesurfer and then did recon-all. After all of that I used 5ttgen hsvs algorithm.
It worked much much more better but still there are problems.
I think manual segmentation is only option for these massive glioma patients