Hi everyone,
I am currently trying to align dwi data and MNI atlas together in datasets. I was using the script below:
#Works without Freesurfer if atlas is taken from the github link below. --eg
#----------------- Convert data to .mif format, denoise, and preprocess
mrconvert $PA_DWI raw_dwi.mif -fslgrad $PA_BVEC $PA_BVAL
dwidenoise raw_dwi.mif dwi_den.mif -noise noise_AP.mif
mrdegibbs dwi_den.mif dwi_den_unr.mif -axes 0,2
dwiextract dwi_den_unr.mif - -bzero | mrmath - mean mean_b0_PA.mif -axis 3
dwifslpreproc dwi_den_unr.mif dwi_preproc.mif -nocleanup -pe_dir PA -rpe_none -eddy_options " --slm=linear --data_is_shelled"
dwibiascorrect ants dwi_preproc.mif dwi_preproc_unbiased_beforemtnorm.mif -bias bias_PA.mif
dwi2mask dwi_preproc_unbiased_beforemtnorm.mif mask.mif
dwinormalise individual dwi_preproc_unbiased_beforemtnorm.mif mask.mif dwi_preproc_unbiased.mif -fslgrad $PA_BVEC $PA_BVAL
dwi2response tournier dwi_preproc_unbiased.mif response.txt
dwi2fod csd dwi_preproc_unbiased.mif response.txt fod.mif -mask mask.mif
mrgrid dwi_preproc_unbiased.mif regrid dwi_upsampled.mif -voxel 2
dwiextract dwi_upsampled.mif - -bzero | mrmath - mean mean_b0_processed.mif -axis 3
mrconvert mean_b0_processed.mif mean_b0_processed.nii.gz
#----------------- Create a WM boundary for seed analysis
#mrconvert $ANAT anat.mif - not necessary
5ttgen fsl $ANAT 5tt_nocoreg.mif
mrconvert 5tt_nocoreg.mif 5tt_nocoreg.nii.gz
fslroi 5tt_nocoreg.nii.gz 5tt_vol0.nii.gz 0 1
flirt -in mean_b0_processed.nii.gz -ref 5tt_vol0.nii.gz -interp nearestneighbour -dof 6 -omat diff2struct_fsl.mat
transformconvert diff2struct_fsl.mat mean_b0_processed.nii.gz 5tt_nocoreg.nii.gz flirt_import diff2struct_mrtrix.txt
mrtransform 5tt_nocoreg.mif -linear diff2struct_mrtrix.txt -inverse 5tt_coreg.mif
flirt -in mean_b0_processed.nii.gz -ref MNI_Glasser_HCP_v1.0.nii -interp nearestneighbour -dof 6 -omat diff2struct_fsl.mat
transformconvert diff2struct_fsl_MNI.mat mean_b0_processed.nii.gz MNI_Glasser_HCP_v1.0.nii flirt_import diff2struct_mrtrix.txt
5tt2gmwmi 5tt_coreg.mif gmwmSeed_coreg.mif
rm hcpmmp1_parcels_coreg.mif
mrtransform MNI_Glasser_HCP_v1.0.nii –linear diff2struct_mrtrix_.txt –inverse –datatype uint32 hcpmmp1_parcels_coreg.mif
#----------------- Run the streamlines
tckgen -act 5tt_coreg.mif -backtrack -seed_gmwmi gmwmSeed_coreg.mif -nthreads 8 -maxlength 250 -cutoff 0.06 -select 10000000 fod.mif tracks_10M.tck #NOT fod_norm - eg
tcksift2 -act 5tt_coreg.mif -out_mu sift_mu.txt -out_coeffs sift_coeffs.txt -nthreads 8 tracks_10M.tck fod.mif sift_1M.txt
tck2connectome -assignment_end_voxels -symmetric -zero_diagonal -scale_invnodevol -tck_weights_in sift_1M.txt tracks_10M.tck hcpmmp1_parcels_coreg.mif hcpmmp_parcels_connectome.csv -out_assignment assignments_sub_hcpmmp1.csv
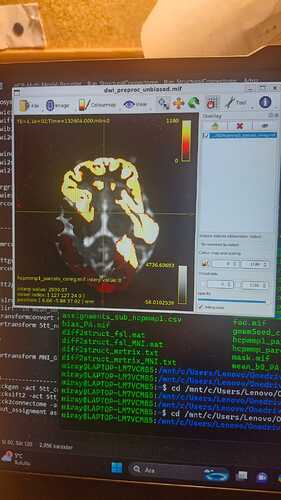
The problem is that it has aligned accordingly in some datasets but failed in others and did not align. In failing alignments I checked 5tt coreg and nocoreg as well as gwmm seed files but they are all adjusted without problem. In failed adjustments the image is like this :
I belive there is a problem in the flirt commands but I could not figure out why and how to fix it.