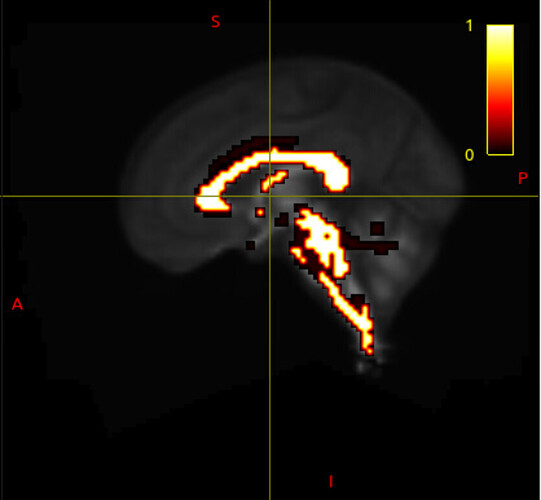
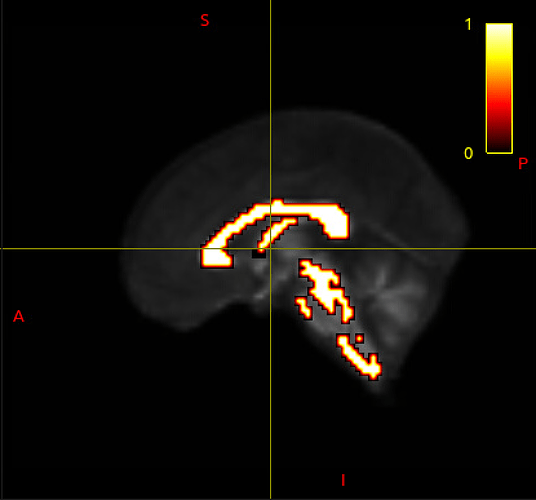
Hi MRTrix forum, I have a rather baffling issue at the moment.
We recently converted our FBA pipeline from v3.0_rc3 to v3.0.2 to take advantage of the improved GLM capabilities in fixelcfestats. (While we were at it, we also added some other niceties like bash parallelization.) Rerunning from the raw data, we get essentially equivalent results at each stage, accounting for a couple of non-deterministic steps in the process, however, when it’s time to actually run fixelcfestats, things fall apart.
Specifically, the uncorrected_pvalue.mif and fwe_1mpvalue.mif files come back with nothing but zeros:
$ mrstats fwe_1mpvalue.mif
volume mean median std min max count
[ 0 ] 0 0 0 0 0 164577
The tvalue.mif and Zstat.mif values also come back as all zeros. After re-checking each step of the process, sometimes redoing steps by hand to ensure that there wasn’t simply a flaw in the script, I found a couple of odd things that I hope will help diagnose the problem:
- For some reason, after the
dwinormalisestep,fa_template.mifandfa_template_wm_mask.mifhave different numbers of voxels:
$ mrstats fa_template.mif
volume mean median std min max count
[ 0 ] 0.0381717 0 0.104609 -0.0280504 0.908895 782582
$ mrstats fa_template_wm_mask.mif
volume mean median std min max count
[ 0 ] 0.0196256 0 0.13871 0 1 973370
This did not happen with the old pipeline:
$ mrstats fa_template_old.mif
volume mean median std min max count
[ 0 ] 0.0325751 0 0.0978714 -0.00965092 0.923676 980796
$ mrstats fa_template_wm_mask_old.mif
volume mean median std min max count
[ 0 ] 0.0209177 0 0.143109 0 1 980796
Despite this, the fa_template and wm_mask do seem to overlay correctly in mrview, so this could simply be a quirk of a change from v3.0_rc3 to v3.0.2. The newer pipeline did give me marginally more brainstem, but that seems deeply unlikely to be causing this kind of problem.
- The
null_contributions.miffiles come back with a single voxel (or fixel, I guess) of 5000 (i.e., the default number of permutations.)
$ mrstats null_contributions.mif
volume mean median std min max count
[ 0 ] 0.0303809 0 12.325 0 5000 164577
$ mrstats null_contributions.mif -ignorezero
volume mean median std min max count
[ 0 ] 5000 5000 N/A 5000 5000 1
Again, this did not happen in the old pipeline:
$ mrstats null_contributions_old.mif
volume mean median std min max count
[ 0 ] 0.0307365 0 0.212712 0 21 162673
$ mrstats null_contributions_old.mif -ignorezero
volume mean median std min max count
[ 0 ] 1.16144 1 0.629731 1 21 4305
This seems potentally more indicative of…something, but I am unable to understand what. Does anyone here have any advice or insights? I feel like there’s something obvious I’m missing but after spending several days trying to track down the problem myself, I’m stumped.
Edit: Another thing that occurred to me is that the stats seem to run much faster on the new pipeline than the old. Would that correspond to whatever process is giving me all zeros on everything? It seems reasonable.