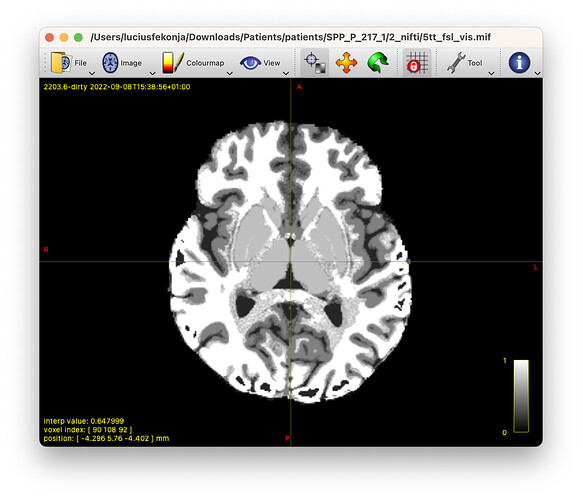
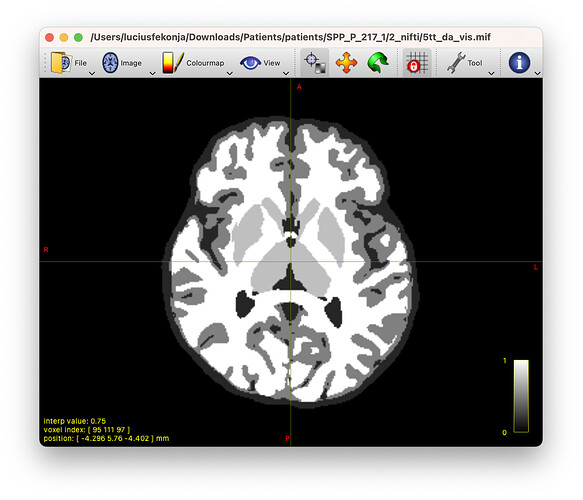
Hi, has anyone used the ANTs’ atropos output for 5ttgen, or has anyone already programmed a possible use (e.g. instead of 5ttgen’s FSL or freesurfer output usage)? Or if not - why? Best, Lucius
FYI, there is now an option available via lib/mrtrix3/_5ttgen/deep_atropos.py under the linked fork.
I have made a simple addition to the 5ttgen script to add a new option via deep_atropos.
This change allows the creation of a 5tt image using antspynet’s ‘antspynet.deep_atropos’ segmentation.
The underlying idea was to allow the generation of a 5tt image based on atropos segmentation, as the results via ‘5ttgen fsl’ gave poor results for T1 images of tumour patients (as well as via ‘5ttgen freesurfer’, and associated ‘5ttgen hsvs’ due to the lesions and resulting intensity changes, or incorrect GM assignment due to the lesions’ contrasts).
The changes include new tissue intensity mappings in a deep atropos segmentation ‘deep_atropos.nii.gz’ where 1=CSF, 2=GM, 3=WM, 4=SCGM, 5=BS and 6=CER. Also, BS and CER are considered WM here due to the nature of deep atropos segmentation.
However, I would welcome further testing and review by others.
I hope this option will be useful to someone else.
Kind regards,
Lucius
edit: added two screenshots to highlight the issues and results.