Hi there experts!
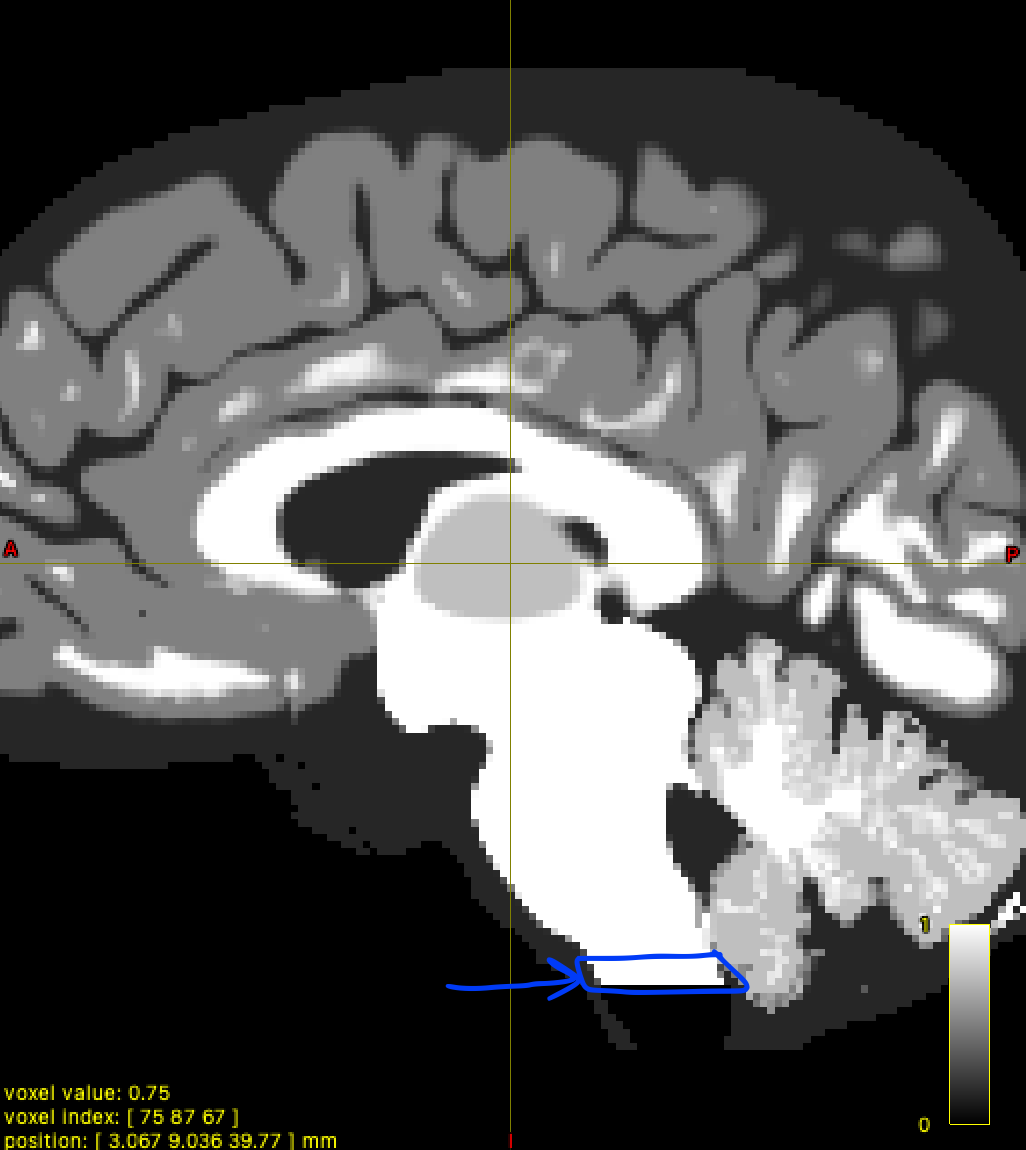
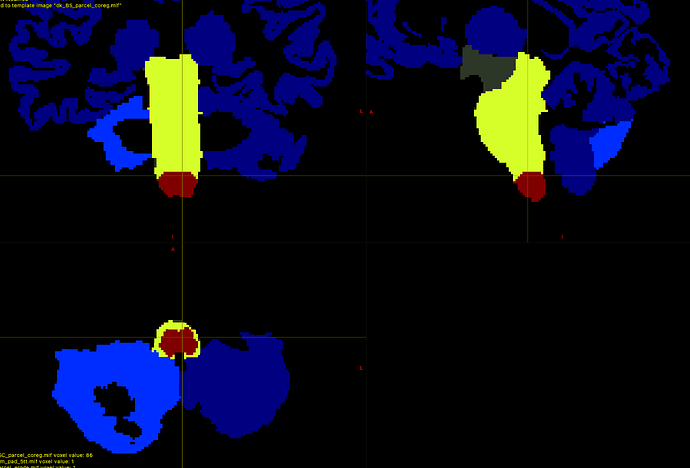
I’m performing anatomically constrained tractography with 5 tissue type images generated by 5ttgen hsvs. I have come up with a way to add 2 ROIs to the parcellation scheme: the brainstem and the spinal cord (the bottom few voxels of the brainstem).
My method seems to do the job but I was wondering if anyone could help me make my method more robust.
labelconvert $fs_dir/mri/aparc+aseg.mgz $FREESURFER_HOME/FreeSurferColorLUT.txt .../fs_default_brainstem.txt dk_BS_parcel.mif # Using a custom LUT to add brainstem to DK parcellation.
mrtransform dk_BS_parcel.mif -linear T1_rig_dwi.txt -datatype uint32 dk_BS_parcel_coreg.mif # Transforming parcellation image to DWI
5tt2vis T1_5tt_coreg.mif 5ttvis.mif
mrthreshold 5ttvis.mif -abs 2 - | mrgrid - regrid -template dk_BS_parcel_coreg.mif brainstem_pad_5tt.mif -datatype bit # Mask of brain stem included in 5ttgen image.
mrthreshold dk_BS_parcel_coreg.mif -abs 85 - | maskfilter - erode -npass 2 brain_parcel_erode.mif # Mask of brainstem from FreeSurfer eroded to make all voxels are within brainstem_pad_5tt.mif
mrcalc brain_parcel_erode.mif brainstem_pad_5tt.mif -subtract brain_parcel_erode.mif -mul - -datatype bit | maskfilter - dilate -npass 5 spinal_cord_dilate.mif # Mask of brain stem not included in 5ttgen image, dilated to include bottom portion of brain-stem segmentation.
mrcalc dk_BS_parcel_coreg.mif 85 -eq spinal_cord_dilate.mif -mul 1 86 -replace dk_BS_parcel_coreg.mif -max dk_BS_SC_parcel_coreg.mif # Adding the bottom portion of brain-stem segmentation back to parcellation image.
For those interested, this paper Connectomes from streamlines tractography: Assigning streamlines to brain parcellations is not trivial but highly consequential - PubMed by Chun-Hung Yeh , Robert E. Smith, Thijs Dhollander, Fernando Calamante, and Alan Connelly gave me the idea.
Many thanks!
Ed