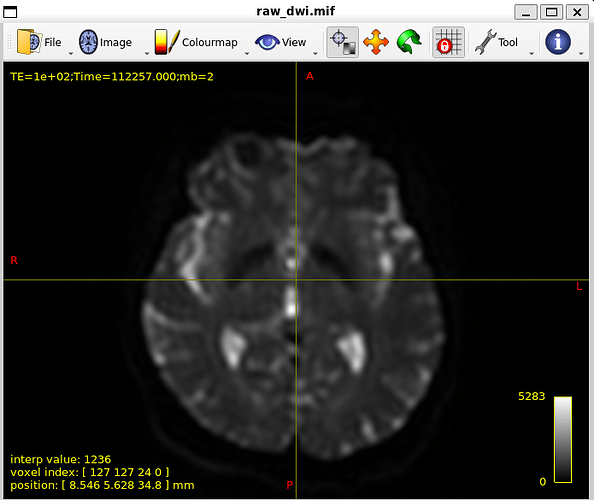
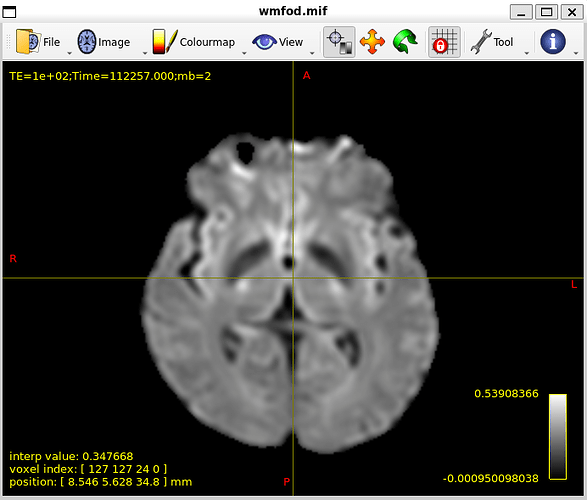
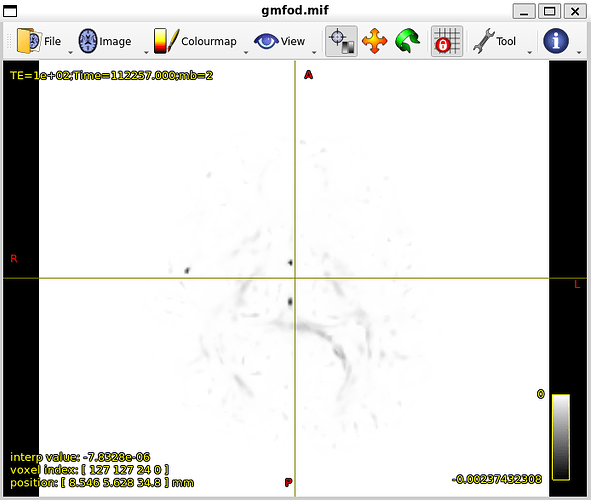
In my dataset whenever I try to create fiber orientation densities whether I use tournier or dhollander I dont get good results.
Is there a problem with preprocessing steps? What should I do in this situation
Preprocessing steps
mrconvert $PA_DWI raw_dwi.mif -fslgrad $PA_BVEC $PA_BVAL
dwidenoise raw_dwi.mif dwi_den.mif -noise noise_AP.mif
mrdegibbs dwi_den.mif dwi_den_unr.mif -axes 0,2
dwiextract dwi_den_unr.mif - -bzero | mrmath - mean mean_b0_PA.mif -axis 3
dwifslpreproc dwi_den_unr.mif dwi_preproc.mif -nocleanup -pe_dir PA -rpe_none -eddy_options " --slm=linear --data_is_shelled"
dwibiascorrect ants dwi_preproc.mif dwi_preproc_unbiased.mif -bias bias_PA.mif
dwi2mask dwi_preproc_unbiased.mif mask.mif
after these steps I do dwi2response and dwi2fod but unfortunately results are not good.
my Raw DWI
my WM-FOD
my GM-FOD