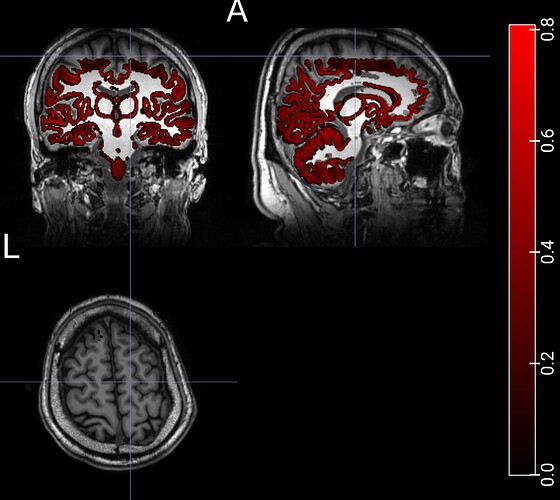
Hello MRtrix3 Community,
I am currently working on creating structural connectomes using anatomically constrained tractography.
For this purpose, I have been utilizing ‘5ttgen fsl’ to generate tractography seeds.
However, I’ve encountered a significant issue: in about 10% of my dataset, which includes almost 100 subjects, the seeds do not seem appropriate, specifically, in the parietal region of the brain.
These are examples of a poor seed and tractography.
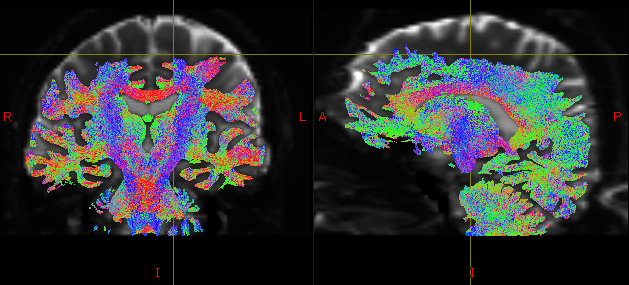
I suspect this problem may be due to the poor quality of white and gray matter segmentation.
(And I guess that could be partly due to inhomogeneities in the T1 signal intensity.)
These are part of the result of segmentation(GM and WM).
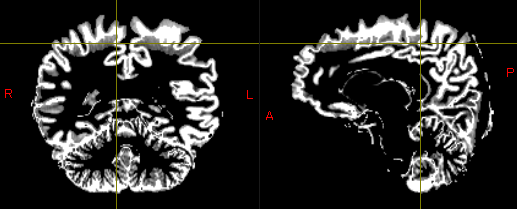
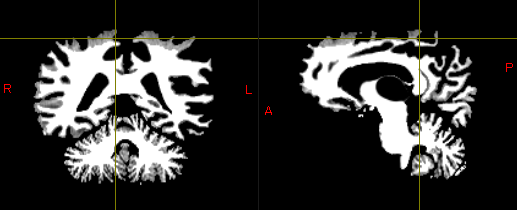
While one potential solution is to redo the process using ‘5ttgen freesurfer’ from the beginning, this approach seems labor-intensive.
Additionally, it feels wasteful to discard the successful results in the remaining about 90% of cases.
I am reaching out to ask if there are any preprocessing strategies or other techniques that could improve seed generation using ‘5ttgen fsl’. Any insights or suggestions to address this challenge would be greatly appreciated.
Thank you in advance!
Best regards,
Kenbridge6