After reading this thread, I tried warping some HCP track files to MNI using the following commands:
warpconvert standard2acpc_dc.nii.gz displacement2deformation warp_subj2MNI.nii.gz
tcktransform 500K.tck warp_subj2MNI.nii.gz 500K_mni.tck
tckmap 500K_mni.tck 500K_mni_tdi.nii.gz -template T1w_restore.nii.gz
fslmaths T1w_restore.nii.gz -mul 0 add 500K_mni_tdi.nii.gz 500K_mni_tdi.nii.gz
(note: last line needed because tckmap doesn’t create the proper nifti headers)
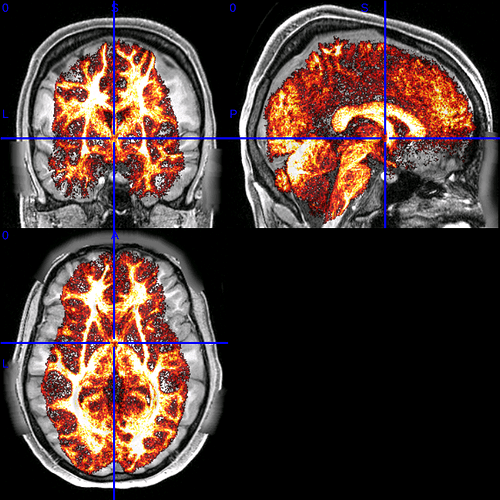
The resulting tdi file looks like this:
You can see that it’s squished, but only on the x axis! I saw in this thread that there was a possible ambiguity with mm vs voxel offsets requiring explicit scaling, but I don’t understand how this can affect only 1 axis?
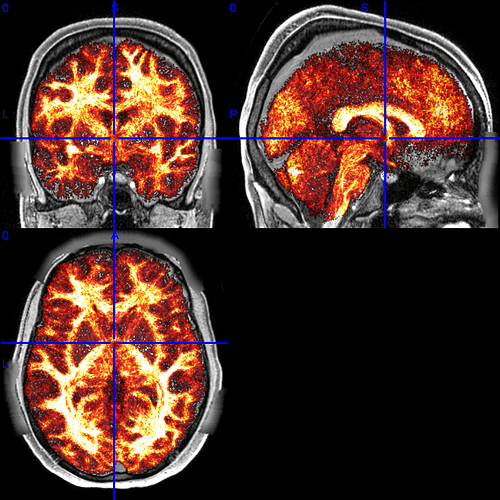
I found that if I mirror the x axis and then scale it by 1.25, the result nearly matches MNI.
mrconvert standard2acpc_dc.nii.gz tmp-[].nii
mv tmp-0.nii x.nii
mrcalc x.nii -neg 1.25 -mult tmp-0.nii
warpconvert tmp-[].nii displacement2deformation warp.nii
tcktransform 500K.tck warp.nii 500K_mni.tck
(including underlay T1 in case it’s possible to toggle between these two and see the subtle shift)
1.25 is the resolution of the diffusion data (though the subject-space T1 image upon which the standard2acpc_dc warp file is based is 0.7mm). However, I still notice a subtle offset, so that combined with the general weirdness of only the x-axis being scaled, and I’m not convinced this is correct.
Any idea what might be going on so that I can be more confident in the output?