I am trying to compute spherical harmonics coefficients for my dwi data after normalising the dMRI data. The sequence is as follows:
$ mrconvert -fslgrad bvecs bvals data_skull.nii.gz data_skull.mif
Here data_skull.nii.gz is the pre-processed HCP dwi data after skull stripping. Next I am extracting the mean b0 and b1000 shells from this multi-shell data.
$ dwiextract -bzero data_skull.mif - | mrmath - mean mean_b0.mif -axis 3
$ dwiextract -shells 1000 data_skull.mif b1000_data.mif
Next I am creating my normalized data:
$ mrcalc b1000_data.mif mean_b0.mif -div normalized_data.mif
Finally to compute SH coefficients I am running:
$ amp2sh normalized_data.mif b1000_coeff.mif
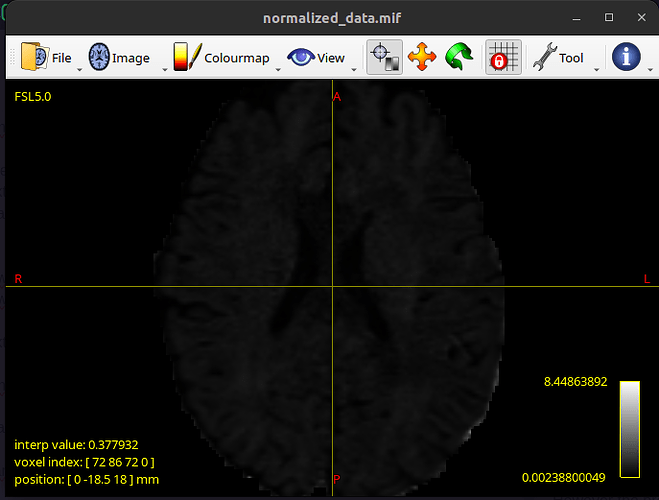
However the problem is with the normalized data. It looks like the following:
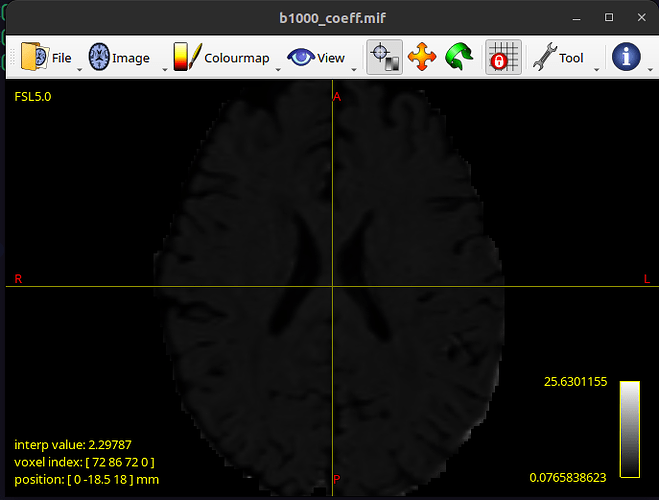
and as a result b1000_coeff.mif looks like this:
Another approach that I tried:
Directly using -normalise option from the amp2sh command. But it requires the b0 image loaded with the b1000 data.
$ mrcat b1000_data.mif mean_b0.mif combined.mif
$ amp2sh combined.mif b1000_coeff.mif -normalise
But doing this gives the error:
amp2sh: [100%] preloading data for "combined.mif"
amp2sh: [ERROR] no valid diffusion gradient table found
amp2sh: [ERROR] error importing diffusion gradient table for image "combined.mif"
As seen above, the original data_skull.mif file had the bvecs and bvals info. Then why it is not reflected here.
What am I doing wrong ? How can I correctly estimate the SH coefficient map ? Any help will be appreciated. Regards.