Hi MRtrixters,
I have a T1 image that has been processed using FreeSurfer (recon-all), followed by labelconvert and labelSGMfix. The T1 image has also been processed using FastSurfer (followed by labelconvert and labelSGMfix).
Both the FreeSurfer parcellation image and the FastSurfer parcellation image look sensible when viewed as the main image in mrview. However, when loading the parcellation image using the -connectome.init option, the FastSurfer nodes are not viewable.
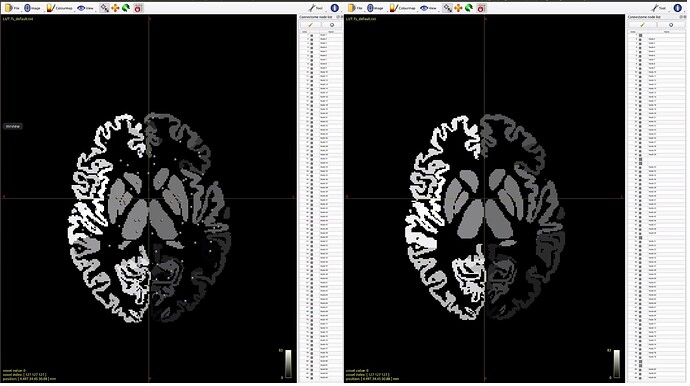
Here is an example, with the left window showing the FreeSurfer parcellation image, right window showing the FastSurfer parcellation image. Both have -connectome.init specified with the corresponding parcellation image.
Is this just a viewing related issue? (and is there a fix?)
Thanks for your help!
Cheers,
Arkiev
Hi @adsouza,
I can’t reproduce this… Any chance you could share the relevant data so I can investigate? Feel free to send the data to my gmail (jdtournier@gmail.com).
Cheers,
Donald
Thanks Donald - you should receive a Cloudstor download link containing example images.
Cheers,
Arkiev
Thanks @adsouza. Looks like this was indeed a bug. I think it works if you change line 2488 of src/gui/mrview/tool/connectome/connectome.cpp as shown on this pull request. You just need to add:
if (nodes[i].get_volume() > 0)
before the node_ordering.insert() call – that seems to fix it on my system.
If you have a source code install, please feel free to give that a try and report back! You just need to edit the file, save it, then issue ./build again and see if that fixed it.
Otherwise, we’ll add the fix to the next release, but it may be a while before we find the time to finalise it… 
1 Like
Thanks you, Donald - I followed the instructions and the nodes are now visible 
Cheers,
Arkiev