Hi @jdtournier ,
Happy New Year.
I hope you are doing great. Thanks a lot for your prompt response always.
I am trying to visualize connectome. I learned that I could get nodes from labelconvert
Mesh could be generated from label2mesh
I am wondering how to get edges and surfaces. How can I get the matrix file for edge visualization? Would you please help me understand these? I would like to get image like this.
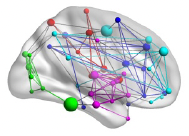
Many Thanks,
Suren
.