Dear MRTrix team,
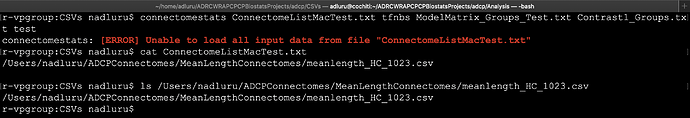
Congratulations on your major release version! I installed the version 3.0.0 that was recently released. I used the git pull approach and built from source on 4/27/2020. I am getting the attached error that I am not able to understand why. The data seems to exist. I was able to load the file in R and also you can see the output of ls in the screenshot. I tried to remove any carriage returns from the input text files and also tried by adding an empty new line but still getting the same error. Just to test things I only placed one file in the list but still getting the same error.
Could you please help me with this?
Thank you so much!
Best,
Nagesh
Hi Nagesh,
I suspect that the error message is a little bit of a red herring here. When it says:
Unable to load all input data from file
, I think that should be read as:
“Unable to load all of the rows within this text file as filesystem paths corresponding to files containing two-dimensional numerical matrix data”.
Given your subjects text file contains the path of a single file, I’m going to have a guess that you’ve concatenated your entire cohort data into a single large 3D matrix, and are now trying to provide that as a single input file. The connectomestats interface is not compatible with this; what it requires (as with the other statistical inference commands in MRtrix3) is a text file for which each row is the filesystem path of a file that contains the data for one “input subject”. So, you need one data file for each subject, where each file contains the 2D connectome matrix data for one specific subject; the input “subjects” text file is then a list of the names of these files, in the same order as the rows of the design matrix.
Cheers
Rob
Hi Rob,
Actually I did not concatenate the cohort data. The reason I had only one path is to try and isolate the reason for the error message. Initially I had 68 rows and it was failing with the same error message, so I wanted to have a single 2D connectome file to check. One thing that occurred to me was that it could be because that the connectome file was created by tck2connectome from an earlier version than 3.0.0. and on a different operating system (CentOS7). While I am trying to do run the connectomestats on MacOS Catalina and Scientific Linux 7. I will try to recreate the connectome file with the current version of it and the same operating system and test again. I just tried even removing the carriage returns from the connectome file itself but it didn’t seem to help.
Thanks so much for your help!
Best,
Nagesh
Okay, looks like I tried to infer too many steps ahead then! 
One thing you could try is loading that one connectome matrix file through the new connectomeedit command. If it fails for the same reason (whatever that may be), that command may provide a more meaningful error message: the code for loading subject data in to connectomestats is 90% shared with e.g. fixelcfestats, so when an error occurs deep within the matrix text file loading code, any useful error message content is vanishing as the error propagates up the code hierarchy.
Hi Rob,
I tried the following.
-
connectomestats with the updated the connectome file by generating it from the latest tck2connectome. Still giving the same error.
-
connectomeedit with to_symmetric on both the old connectome and the updated connectome file and the command worked fine with both of them.
Since it is not obvious to me what I should try next, I am providing the connectome file and sample design and contrast matrices. The design matrix is a dummy one. Hope you could take a closer look this way! Please also let me know if I need to try any additional things on my end.
Connectome
Design
Contrast
Textfile for connectomestats
The actual command for connectomestats
connectomestats ConnectomeListMacTest3.txt tfnbs ModelMatrix_Groups_Test.txt Contrast1_Groups.txt test
Thanks so much for all your help, Rob!
Best,
Nagesh
Hi Nagesh,
Thanks for sharing the data, and congratulations on reporting the first bug post-3.0.0! 

Without boring everyone with the details, you can get the command to work by, when providing connectomestats with the path of the file that contains the subject list, put “./” at the start of the file path to indicate that the location of that file is the current working directory:
connectomestats ./ConnectomeListMacTest3.txt tfnbs ModelMatrix_Groups_Test.txt Contrast1_Groups.txt test
For readers, this should not affect fixelcfestats, since in that case a fixel directory is provided as a “reference location” from which to find those subject files. A fix will be pushed out nonetheless.
Rob
2 Likes
Thanks so much, Rob! It works now.